About Authors:
Vaibhav Patel*, Punit Bhatnagar, Gopal Rai, Alok Pal Jain
Guru Ramdas Khalsa Institute of Science & Technology (Pharmacy),
Jabalpur
*vaibhavpatel281@gmail.com
Introduction
Gene therapy by small interfering RNAs (siRNAs) hasbeen emerging as innovative nucleic acid medicines with increasing knowledge on the molecular mechanisms of endogenous RNA interference. Gene silencingis a promising tool for the treatment of numerous human diseases that cannot be cured by rational therapies. The primary success of siRNA applications depends on suitable vectors to deliver therapeutic genes.
Reference Id: PHARMATUTOR-ART-1386
Historical Aspect
siRNA was first discovered by David Baulcombe’s group at the Sainsbury laboratory in Norwich, England as a part of post-transcriptional gene silencing (PTGS) in plants. This innovative output was first published in a paper entitled as “A species of small antisense RNA in post-transcriptional gene silencing in plants” in October 1999. In the same sequence various studies has been continuously carried out to explore the therapeutic potential of siRNA to its utmost extent.
siRNAalso known as short interfering RNA or silencing RNA. It is a class of double-stranded RNA molecules of 20-25 nucleotides in length having a 5' phosphate group and a 3' hydroxyl (OH) group, with 3' overhangs (2 nucleotides) at each end that can be used to "interfere" with the translation of proteins. They specifically bind to a particular sequence of gene on mRNA and thus by promoting the degradation of specific proteins they act as a biotechnology tool to treat diseases like cancer etc. In this way, they prevent the production of specific proteins based on the nucleotide sequences of their corresponding mRNA. The process is called RNA interference (RNAi), and may also be referred to as “siRNA silencing or siRNA knockdown”. si-RNA is generally considered to have come from longer strands of exogenous RNA which is taken up by the cell and undergoes further processing.
The siRNA can be successfully delivered by novel drug delivery tools like nanoparticles, liposomes, micelles, vectors, viruses or transposons etc. Once they entered the cell they are acted upon by RNase III–like enzyme, called Dicer, using restriction enzymes. The si-RNA is then incorporated into a multi-subunit protein complex called RNAi induced silencing complex (RISC). RISC "seeks out" an appropriate target mRNA. In the same sequence it was hypothesized by some researchers that; it acts as an antisense strand which directs the degradation of the complimentary strand of mRNA, by using a combination of endo- and exonuclease enzymes. siRNA have been found to play a crucial role in antiviral defense, degradation of over-produced mRNA, in shaping the chromatin structure of a genome or preventing disruption of genomic DNA by transposons. Many diseases can potentially be treated by inhibiting gene expression due to which siRNA has drawn a major attention of various academic researchers and biopharmaceutical companies.
Design for synthetic siRNA
Recently numbers of designs for the manufacturing of siRNA has been put forward by scientists and along with that they are continuously trying to espy some alternate protective drug delivery route for safe and effective delivery of siRNA.
General Guidelines for the Design of siRNA
- siRNA targeted sequence is usually 21 nucleotide in length.
- Avoid regions within 50-100 bp of the start codon and the termination codon.
- Avoid intron regions.
- Avoid stretches of 4 or more bases such as AAAA, CCCC.
- Avoid regions with GC content <30% or > 60%.
- Avoid repeats and low complex sequence.
- Avoid single nucleotide polymorphism (SNP) sites.
- Perform BLAST homology search to avoid off-target effects on other genes or sequences.
- Always design negative controls by scrambling targeted siRNA sequence.
A delineative architecture of a typical hairpin siRNA developed by a siRNA expression vector is described in the given Figure 1.
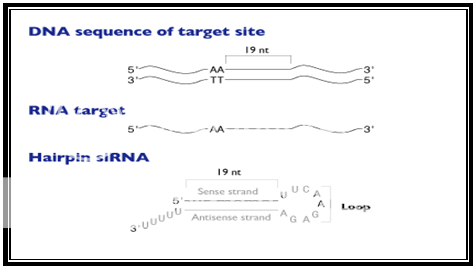
Figure 1.Schematic representation of a typical hairpin siRNA produced by a siRNA expression vector
Synthesis of siRNA
Recommendations for siRNA hairpin design and cloning strategy are made based on research by Ambion scientist's procedures. The first step in designing an appropriate insert is to choose the siRNA target site which is done by scanning an mRNA sequence for AA dinucleotides and recording the 19 nucleotides immediately downstream of the AA. After that a siRNA expression cassette is constructed in a specific order of sense strand, short spacer and antisense strand. The length of the siRNA stem should range between 19 to 22 nucleotides-long sequences and the loop size should lie between 3 to 23 nucleotides. Various scientific studies have revealed that these specific stem lengths and loop sizes functions well in gene silencing studiess
Techniques of siRNA delivery
I. Transfection – In this technique, siRNA are complexed with a suitable carrier such as liposome that allows them to traverse through cell membranes. This technique has high efficiency ad reproducibility in most cell types. Transfection typically involves the use of a wide variety of carriers to facilitate the efficient cellular uptake of siRNA. Some major carriers used for this technique are liposomes, polymer based nanoparticles, dendrimers, polymer peptides, aptamers, micelles, etc.
II. Electroporation– In electroporation technique, a brief but powerful electric pulse is applied due to which the lipid molecules reorient and undergo thermal phase transition due to heating resulting in the transient creation of hydrophilic pores in the cell membranes. The pore formation results in uptake of siRNA and drug moiety by the cell. Despite of high efficiency of nucleic acid transfer, electroporation induce high cell mortality.
III. Viral vector– Delivery of siRNA from DNA template can be carried out by several recombinant viral vectors based on retrovirus, adeno associated virus, adenovirus ad lentivirus. These viral vectors have been engineered and optimized to facilitate the entry of siRNA into cells that are not readily amendable to transfection. This technique has high efficiency for in-vivo delivery into primary and non dividing cells.
Applications of siRNA Technology
An advanced field of applications had been arisen from siRNA technology which is achieving a humongous space among the traditional therapeutic system. Some of the customary applications of the siRNA therapy are exemplified in the Figure 2.
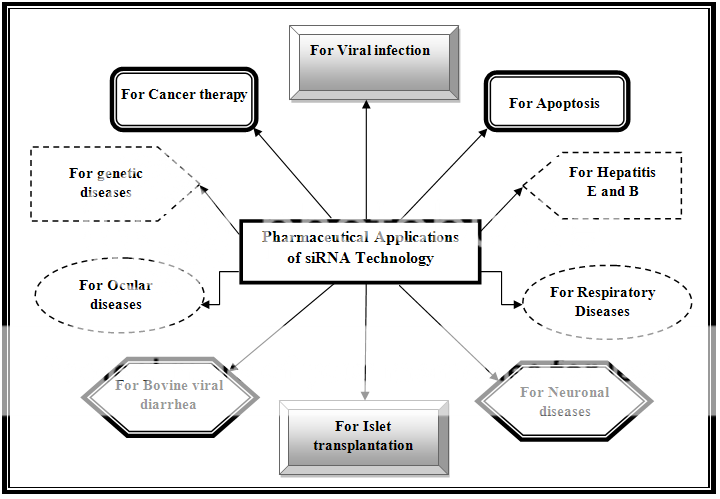
Figure 2:Various applications of siRNA Technology
Challenges of siRNA therapy
Although siRNAs offer several leading edge advantages over existing conventional therapies, but still it is striving with potential challenges which is to be overcome in near future. One of such threatening challenge is therapy for an ‘off-target’ effect which is due to inhibition of a gene, the expression of which should not be targeted, because the gene shares partial homology with the siRNA. The inadvertent silencing of non-target genes may lead to problems in interpretation of data and potential toxicity. To avoid this issue, the design and selection of potent siRNAs should be carefully performed.
Another challenge facing siRNA therapy is ‘immune stimulation’; this is recognition of a siRNA duplex by the innate immune system. Introduction of excess of siRNA is known to result in nonspecific events owing to activation of innate immune responses. The sequence issue of siRNA-mediated immune stimulation still needs to be investigated.
A very crucial challenge in siRNA therapy is the issue of delivery. Being an RNA entity, siRNA is anionic, hydrophilic, and unable to enter cells by passive diffusion mechanisms. Moreover, in-vivo delivery of naked siRNA to appropriate disease sites remains a considerable hurdle owing to rapid enzymatic digestion in plasma and renal elimination, limited penetration across the capillary endothelium, and inefficient uptake by tissue cells. To overcome these difficulties, the development of effective in vivo delivery systems is essential.
Future Prospects
siRNA therapeutics has several distinct advantages over traditional pharmaceutical drugs. RNAi is an endogenous biological process, so almost all genes can be potently suppressed by siRNA. siRNA-based therapeutics offer an enchanting and sovereignpath to treat multiple cancers because the RNAi pathway enables the specific silencing of pathogenic genes involved in cancer progression. Analysis methods that can conveniently detect siRNA in biological samples should be concurrently developed. Effective and optimized delivery systems for therapeutic siRNA will make it easier to access cancer therapy and provide good cancer therapeutic models. The efficacy and specificity of cancer treatment will be further enhanced by using a combination approach of siRNA with another traditional therapy such as chemotherapy, radiotherapy, photodynamic therapy, or immunotherapy.
The development of effective and safe delivery systems for therapeutic siRNA is crucial. Besides the development of viral vectors as delivery vehicles, an increasing number of highly diverse nonviral systems are evolving.
References
1. S.M. Elbashir, J. Harborth,W. Lendeckel, A. Yalcin, K.Weber, T. Tuschl, Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells, Nature 411 (2001) 494–498.
2. G. Meister, M. Landthaler, A. Patkaniowska, Y. Dorsett, G. Teng, T. Tuschl, Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs, Mol. Cell. 15 (2004) 185–197.
3. J.S. Parker, S.M. Roe, D. Barford, Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity, EMBO J. 23 (2004) 4727–4737.
4. C.D. Novina, M.F. Murray, D.M. Dykxhoorn, P.J. Beresford, J. Riess, S.K. Lee, R.G.
5. Collman, J. Lieberman, P. Shankar, P.A. Sharp, siRNA-directed inhibition of HIV-1 infection, Nat. Med. 8 (2002) 681–686.
6. E. Song, S.K. Lee, J. Wang, N. Ince, N. Ouyang, J. Min, J. Chen, P. Shankar, J. Lieberman, RNA interference targeting Fas protects mice from fulminant hepatitis, Nat. Med. 9 (2003) 347–351.
7. E. Song, S.K. Lee, J. Wang, N. Ince, N. Ouyang, J. Min, J. Chen, P. Shankar, J. Lieberman, RNA interference targeting Fas protects mice from fulminant hepatitis, Nat. Med. 9 (2003) 347–351.
8. M.A. Behlke, Progress towards in vivo use of siRNAs, Mol. Ther. 13 (2006) 644–670.
9. Melnikova, RNA-based therapies, Nat. Rev. Drug. Dis. 6 (2007) 863–864.
10. J.T. Marques, B.R. Williams, Activation of the mammalian immune system by siRNAs, Nat. Biotechnol. 23 (2005) 1399–1405.
11. R. Juliano, M.R. Alam, V. Dixit, H. Kang, Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides, Nucleic Acids Res. 36 (2008) 4158–4171.
12. Aigner, Nonviral in vivo delivery of therapeutic small interfering RNAs, Curr. Opin. Mol. Ther. 9 (2007) 345–352.
13. D.A. Braasch, S. Jensen, Y. Liu, K. Kaur, K. Arar, M.A. White, D.R. Corey, RNA interference in mammalian cells by chemically modified RNA, Biochemistry, 42 (2003) 7967–7975.
14. H.R. Kim, I.K. Kim, K.H. Bae, S.H. Lee, Y. Lee, T.G. Park, Cationic solid lipid nanoparticles reconstituted from low density lipoprotein components for delivery of siRNA, Mol. Pharm. 5 (2008) 622–631.
15. S.I. Kim, D. Shin, T.H. Choi, J.C. Lee, G.J. Cheon, K.Y. Kim, M. Park, M. Kim, Systemic and specific delivery of small interfering RNAs to the liver mediated by apolipoprotein A-I, Mol. Ther. 15 (2007) 1145–1152.
16. D. Peer, E.J. Park, Y. Morishita, C.V. Carman, M. Shimaoka, Systemic leukocyte directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target, Science, 319 (2008) 627–630.
17. S.D. Li, Y.C. Chen, M.J. Hackett, L. Huang, Tumor-targeted delivery of siRNA by self assembled nanoparticles, Mol. Ther. 16 (2008) 163–169.
18. S.I. Pai, Y.Y. Lin, B. Macaes, A. Meneshians, C.F. Hung, T.C. Wu, Prospects of RNA interference therapy for cancer, Gene Ther. 13 (2006) 464–477.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to PharmaTutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE









