{ DOWNLOAD AS PDF }
ABOUT AUHTOR
Nitisha Bhandari*, Akanksha Bhandari
Shri Guru Ram Rai Institute of Technology and Science, Patel Nagar,
Dehradun 2Graphic Era University, Dehradun, India
*nitishabhandari89@gmail.com
ABSTRACT
Interactions of proteins could be resulted from homodimerisation or heterodimerisation. But these are quite specific and always selective in nature and act onlyon particular set of class of proteins, which provide substantial target behaviour to a drug designer. Here in this paper, provided a commentary based review compiling the short notes information provided on protein proteins interactions, by “Catherine Royer”
[adsense:336x280:8701650588]
REFERENCE ID: PHARMATUTOR-ART-2472
|
PharmaTutor (ISSN: 2347 - 7881) Volume 5, Issue 3 Received On: 19/10/2016; Accepted On: 24/11/2016; Published On: 01/03/2017 How to cite this article: Bhandari A, Bhandari N;Protein-protein surface interactions: Constrains of homologous versus heterogeneous domains; PharmaTutor; 2017; 5(3); 9-14 |
INTRODUCTIONS
Interaction of biomolecules lead to the various biochemical events. Even drugs derived from natural source, as well as synthetics, also needs to interact with biomolecules to affect their activity[1-3].Althoughprotein-protein interactions (PPIs) are the keys, toregulate or transduce a signal to a function for maintaining a normal homeostasis, but any fault leads to a malfunctioning and diseases (say e.g. anaphylactic shock)[4, 5].Around > 99% metabolic routes utilised the PPIs for tight hold. In recent years, various genomic tools and techniques[6, 7] were used to understand the nature of PPIs and now it is much established that these interactionscan often result from aphysical contacts, involving bonding and electrostatic forces.
PPIs are critical for intracellular signalling, endocytosis and internalisations. They are crucial to the understanding of all in vivo functions, cellular regulation, biosynthetic and degradation pathways, signal transduction, Nucleic acid replication, transcription and translation, multi-molecular assembly associations, cellular packaging, optimisation of immune response. They also relate to allosteric mechanisms, either turning genes on or off and therefore attain high interest for drug designing. All biological processes are regulated through association and dissociation of protein molecules. These includesendogenous biomolecules receptor binding, protease inhibition of protein lysis, Antibody-antigen recognition efficacy, signal transduction mechanisms, enzymesubstrate affinity, vesicle transport, Post-translations or -transcriptions modifications. Hence, PPIs are now highly targeted and become centre stage of protein science. The ability to predict the preferred method by which proteins interact would facilitate assignment of protein function.
Classes of Protein-Protein Interactions
Majorly PPIs resulted intothe complexes formation, as homo-oligomers orhetero-oligomers. The homo-oligomerare macromolecular complexes constituted by only one type of protein subunit. Protein subunit assembly is guided by the establishment of non-covalent interactions in the quaternary structure of the protein. Disruption of homo-oligomers in order to return to the initial individual monomers often requires denaturation of the Complex. Several enzymes, carrierproteins, scaffold proteins, and transcriptional regulatory factors carry out their functions as homo-oligomers. Distinct protein subunit interacts in hetero-oligomers, which are essential to control several cellular functions. The importance of the communication between heterologous protein is even more evident during cell signalling events and such interactions are only possible due to structural domains within the proteins.
There is another category where they are considered their interactions in stability context.Stable interactions involve proteins that interact for a long time, taking part of permanent complexes as subunits, in order to carry out structural or functional roles. These are usually the case of homo-oligomers (e.g. cytochrome c), and some hetero-oligomeric proteins, as the subunit of ATPase. On the other hand, a protein may interact briefly and in a reversible manner with other proteins in only certain cellular contexts-cell type, cell cycle stage, externalfactors, presence of other binding proteins etc. These are called transient interactions(e.g.- Could be seen in 5HT receptor-macromolecular complexes [8]). The another way to classify these interactions as covalent and noncovalent interactions. Covalent interactions are those with the strongest association and are formed bydisulphide bonds or electron-sharing.Although being rare, these interactions are determinant in some posttranslational modifications, as ubiquitination and sumoylation. Noncovalent interactions are usually established during transient interactions by the combination of weaker bonds, such as hydrogen bonds, ionic interactions, van der Waals forces, or hydrophobic bonds.
Homo-oligomerization
The noncovalentinteractions of protein quaternary structure provide, an edge over the cases where thefunctional unit comprised of a linear polypeptide chain. Initial binding of ligand excels energy on subunit site of protein and drives it to adopt an alter transition conformation. However, the modulation of subunit affinity towards the ligand binding is independent of the adopted fold structure of the protein, providing a considerable energetic marginfor modulation of activity. Also, these experiments told the protein regulatory functions works up to an appropriate molar concentration range and later the affinity of protein subunit fades out, indicating the role of protein concentration dependent. These regulatory functions can either activate or inactivate the progression of any cellular process induced by oligomer or the monomer. Hence it is worth to say that the function in oligomers can fine-tuned by available ligand concentration (could any ions,ortho- or allosteric substrate and ligandsetc.) and by protein concentration (comparative level of expression ordegradation rates), see figure as an example of homooligomerisation.
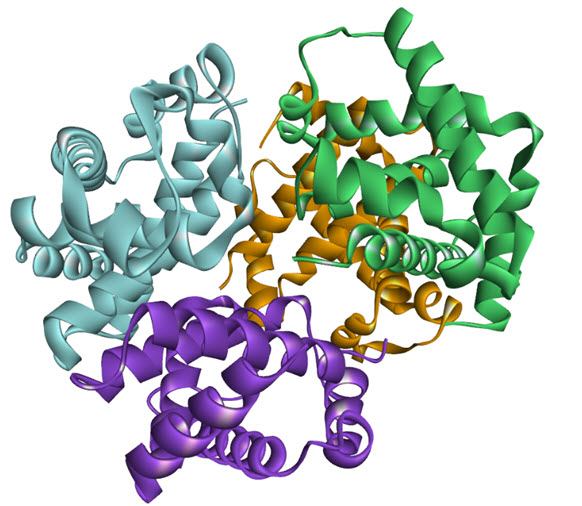
Figure 1. Homo oligomerization of haemoglobin tetramer (PDB: 1GZX)[9]
Interactions between heterologous proteins
More often the heterologous protein interactions came to known and become more popular when it came apparent that physical state or the ambient environmentalaffectsthe cellularmetabolic rate (like these interacting proteins can be affected by thechemical presence, change in luminosityetc). However later math and current research found them more valuable and critical in various other, metabolic and much required homeostasis functions. Although here we encompass below some of the heterologous proteins.
Protein domains involved in Signal Transduction
Src homology domains 2 and 3,are commonly referred to as SH2 and SH3 domains. The SH2domains recognize tyrosine phosphorylated proteins (particular autophosphorylatedgrowth factor receptors)[10]. A ribbon cartoon shown below, indicating the SH2 domain of the lymphocytespecific tyrosine kinase, P56lck along a co-crystallise peptide [11], see figure 2. The structuralarchitecture of SH2 domains possessof a three-stranded twisted beta sheet which is sandwichedbetween two alpha-helices. The SH2 domain also have a deep cleft which acquired high affinity for phosphotyrosine, but not for, either phosphoserine or phosphothreonine.
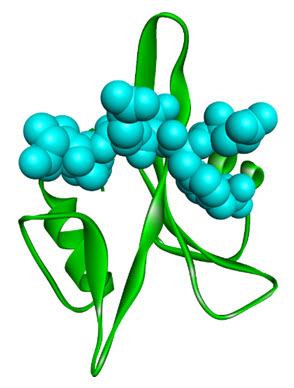
Figure 2. Ribbon diagram of the SH2 domain from p56lck (green ribbons) bound to phosphopeptide (spacefilled cyan) (PDB: 1CWE).
Residues starting for C-terminal up tophosphotyrosine, has been noticed as an active site to conferthe specificity of the interaction. Whereas the second pocket assist in recognizing these C-terminalspecificity determinants. Various classes of SH2 domains recognize numerous typesof C-terminal sequences (Class I SH2 domains recognize C-terminal tripeptidesP-P-H, where P and H stand for polar and hydrophobic; Class III SH2 domainsrecognize H-X-H tripeptides). Till this date, number of SH2 domains three dimensional structures have been resolved and found a clear evident of structural homology (predicted by the comparative analysis of SH2 domain from p561ck andC-terminal SH2 domain of the Syk tyrosine kinase)[12].
Like SH2 domains, SH3 domains are also found in similar signal transducer proteins (protein tyrosine kinases), however they recognize polyproline type II helical structures (PXXP motifs) in their binding proteins(e.g., SH3 domain of Grb2recognizes the PXXP motif of a guanyl nucleotide releasing factor;SH3 domain of the src kinase, Fyn, canbind the PXXP motif of the HIV-1 protein Nef;SH3 domain of fyn proto-oncogene complexed with asynthetic p85 subunit peptide of Phosphoinositide 3-kinase) [13] is shown in Figure 3. It could be easily seen that the SH3 domainarchitecture possesses of a beta barrel formed by two orthogonal beta sheets, which is constructed of threeanti-parallel beta strands and the proline rich site on the partner proteins pack onto aromatic residues of the SH3 domains.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT editor-in-chief@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
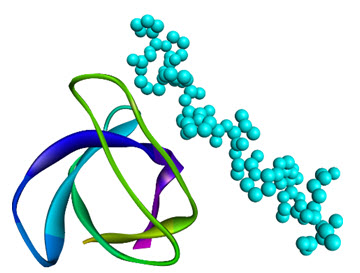
Figure 3.NMR Structure of the SH3 Domain from Fyn Proto-Oncogene Tyrosine Kinase complexed with the synthetic peptide P2l corresponding to residues 91-104 of the P85 Subunit of PI3-Kinase, (PDB 1AZG)
PDZ domains is an another class of protein interaction motifs which has GLFG repeats or DHRdomains and were identified initially in three proteins, PSD-95, DlgA andZO-1 (therefore they called so, “PDZ domains”) where all are guanylate kinases. These are highly involved in ion channel receptor clustering, receptor/enzymecoupling and a variety of other protein assembly associations. The PZD domains recognize C-terminaltri-peptide motifs (S/TXV), other PZD domains or even LIM domains (see in Figure 4).Surprising resemblance of PDZ domain of DglAwith SH2 domain, tells similar structural architecture includinga beta barrel sandwiched between two alpha-helices. The below figure shows the structural PDZ domain of PSD95 holding tightly the C-terminal peptide from Cript[14].
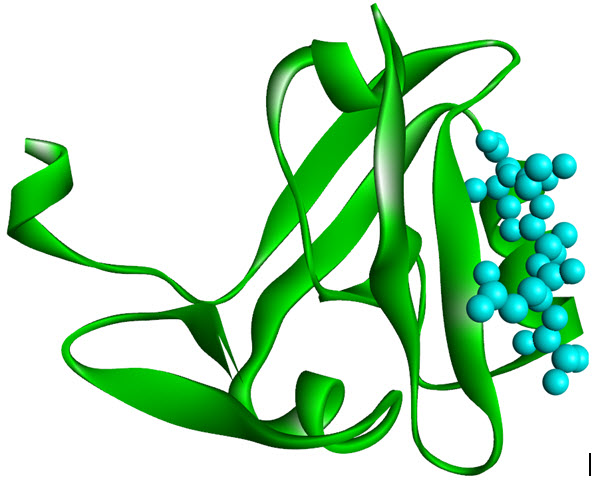
Figure 8. PDZ domain of PSD-95 complexed with the C-terminal peptide derived (PDB 1BE9)
The another cellular signalling domain is the LIM domain. These domains arecysteine enrichedzinc finger domains present in many homeodomain and non-homeodomain proteins involved critically in cellular development,differentiation, association withthe cyto-skeletons or in cellular senescence. These proteins need PDZ motifs, bHLHtranscription factors (see below), other LIM domains, and LIM binding proteins to bind.These were initially found in 3 homeodomain transcription factors (lin11, isl1 and mec3, henceLIM domain), which holds the consensus sequence CX2CX16-23HX2CX2CX2CX16-21CX2C/H/D, andnow identified in over 35 proteins to date. The NMR structureof the LIM domain from the LASP protein was first identified in 1996[15].The zinc atom (in space-filled cyancolour), coordinated by cysteine residues in the two loops corresponding to the double finger, also shown in figure 5.

Figure 5. Structure of the amino terminal of LIM domain peptide of the LASP protein (PDB:1ZFO)
The pleckstrin homologydomain (also called, “PTB domains”) proteins also transduce signal in cellular intracellular signalling. These proteins bind to acidic domainsin signal transducer proteins as well as to phosphoinositdes, the WW domain, a semiconservedregion of 38-40 amino acids, containing two conserved tryptophan at 20 amino acid spaced and hence called soas “WW”(WSxWS motif in cytokine receptors and the WD repeat).
Protein domains involved in transcriptional regulation
The nucleic acid protein juncture has been highlighted since when it gets focused by the researcher for the first time, especially modulation of the gene expression gets favouritism from the researchers in 21st century. Although these processes require selective recruitment of specific proteins,selective energy based conformation for the affinityand hence highly structural specific.Numerous prokaryotic and eukaryotic transcription factors were found by biochemists with or without their key role in the cell. For better understanding, 3D structures were characterised and classified further,basedon sequence or structural homology, except some instances where they found on functional homology.
One of highly focused family is that of the bHLH transcription factors, so called as its structural motif include a basic region for binding DNA and a helix-loop-helix regioninvolved in dimerization. This family include the MyoD family oftranscription factors which regulate the gene expression in muscle cells and control their differentiation to mature muscle cells. Other members of the bHLH family are the ubiquitous products of the E2A gene, E12 and E47. Although MyoD family members can form homo as well as heterodimers with the E12 and E47 proteins, see figure 6.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT editor-in-chief@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
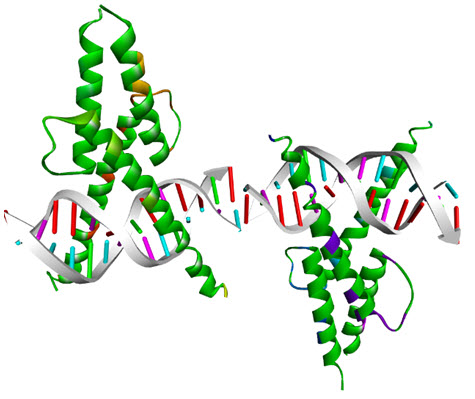
Figure 6.bHLH domain of MyoDcomplexed with DNA (PDB 1MDY)
Another class includesleucine zippertranscription factors,c-jun and c-fos (constituents of the AP1 transcriptionfactor). AP1 is highly involved in gene transcription regulation incell proliferation. Similar to bHLH family,bzip family members also interacts by their coiled alpha helices. Interestingly, the bzip proteins possess the leucine residues positioned athelix turn providing ample hydrophobic interface for the PPIs,hence so called as leucine zipper. N-terminal to the zipper recognizeand bind to specific DNA sequences. These family members also homo- andhetero-dimerize together or other family protein and can alter the DNAspecificity and the transcriptional rate (see in figure: b-zip domains of the c-Fos/cJun heterodimer bound to DNA, see in Figure 7)[16].

Figure 7.Schematicrepresentation of c-Fos/c-Jun heterodimer complexed with DNA (PDB 1FOS)
The RHR or Rel Homology Region containing transcription factors are another class that are functional active in cellular immune responses orfurther de-differentiation. NF-κB, is one of the key member of this class and has specificity to DNA target site in the immunoglobulin light chain enhancerand also regulating various gene expression in response to invading infection. TheRel Homology Region of the two subunits of NF-κB (p50 and p65 or RelA) specifies DNA binding, dimerization and nuclear localization. Crystallographic data revealed RHR region in the p50 subunit of NF-kB[17]resembledlike beta sandwich structure of the immunoglobulin folds ofimmunoglobulins and also assist in the dimerization with DNA. Ankyrin repeatare ubiquitous domainsand found in more than 100 different proteins of diverse functionality like catalytic, transcription or inhibitors of transcription activitiessimilar-like IkB, that inhibits the NF-kB transcription factor. Ankyrin repeats contain around 32-33 amino acids and usually proteins hold at least 4 copiesof this domain. The structural frame processes beta strand, helix turn helix extendedstrand beta strand segment. Whereas the helices and beta strand are anti-parallel to each other and theplane of the strand regions is perpendicular to the helical axis (See Figure 13 below). Theconsensus sequence of the ankyrin repeat is:
- G – T P L H L AA R – G H V E V V K L LL D – G A D V N A – T KA I S Q N N LD IA E V K N P D DV K T M R Q S I NE
Another large family includes the family of steroid/nuclear receptorsincludingsteroidal hormonereceptors, vitamin D receptor, retinoic acidreceptor (RAR and RXR) and thyroid hormone receptors, and a several other orphan receptors andtranscription factors. The structural construction is quite unique of these receptors comprising N-terminal constitutive and tissue specifictranscriptional activation domain (TAF1) which can have varied in length, central region for DNA binding anddimerization domain (DBD, linker region for nuclear localization and C-terminalas ligand binding domain (LBD) that also holds ligand dependent transcriptional activationactivity (TAF2) and dimerization determinants.However, the structural information of the DBD and LBD of these receptorsdrawn interest of putative drug targets, but also challenged the drug designer as they share high degree of structural homology in their structures. The individual DBD homodimerizes with DNA through palindromic response elements (HRE’s) and shows cooperativity relationship for the binding of the other DBD of the protein on DNA.

Figure 8. Structure of the ERDBD bound to DNA
It was also seen that these DNA Binding Domains (DBD)have highly conserved pair of Zn fingers, one of which involves in DNA recognition helix, andthe other holds the determinants for dimerization. However,the D-loop participate in the dimer interface, flanked by two cysteine residues which coordinates with Zn atom (see in Figure 8, showing the DBD from the estrogen receptor (ER) bound to an oligonucleotidebearing the estrogen response element sequence [18].
Acknowledgment
I would like to thanks all the people who helped during the preparation of this manuscript and also like to acknowledge the pioneer work of Catherine Royer (molecular biophysics, Professor in the Department of Biology in the School of Science in Rensselaer Polytechnic Institute)in the field of protein-protein interactions.
References:
1. Negi A, Gill B: Success stories of enolate form of drugs. PharmaTutor 2013, 1(2):45-53.
2. Singla R, Negi A, Singh V: Indole based alkaloid in cancer: an overview. PharmaTutor Mag 2014, 2:76-82.
3. Singla R, Singh V, Negi A: Synthetic Indole Alkaloids in Cancer: An Overview.
4Negi A: Anaphylactic Shock: Shocking Error of Immune System!PharmaTutor 2013, 1(2):12-16.
5. Negi A, Pandey AK, Joshi G, Agnihotri V: Impact of protein tyrosine phosphate on cancer metastasis: an overview. World J Pharm Res Technol 2013, 1(2):118-130.
6. Negi A, Gill B, Anand S: Tilling: versatile reverse genetic tool. PharmaTutor 2014, 2(1):26-32.
7. Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C: STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research 2013, 41(D1):D808-D815.
8. Zhou J, Reidy M, O’Reilly C, Jarikote DV, Negi A, Samali A, Szegezdi E, Murphy PV: Decorated Macrocycles via Ring-Closing Double-Reductive Amination. Identification of an Apoptosis Inducer of Leukemic Cells That at Least Partially Antagonizes a 5-HT2 Receptor. Organic letters 2015, 17(7):1672-1675.
9. Paoli M, Liddington R, Tame J, Wilkinson A, Dodson G: Crystal structure of T state haemoglobin with oxygen bound at all four haems. Journal of molecular biology 1996, 256(4):775-792.
10. Negi A, Ramarao P, Kumar R: Recent Advancements in Small Molecule Inhibitors of Insulin–like Growth Factor-1 Receptor (IGF-1R) Tyrosine Kinase as Anticancer agents. Mini reviews in medicinal chemistry 2013, 13(5):653-681.
11. Tong L, Warren TC, Lukas S, Schembri-King J, Betageri R, Proudfoot JR, Jakes S: Carboxymethyl-phenylalanine as a replacement for phosphotyrosine SH2 domain binding. Journal of Biological Chemistry 1998, 273(32):20238-20242.
12. Narula S, Yuan R, Adams S, Green O, Green J, Philips T, Zydowsky L, Botfield M, Hatada M, Laird E: Solution structure of the C-terminal SH2 domain of the human tyrosine kinase Syk complexed with a phosphotyrosine pentapeptide. Structure 1995, 3(10):1061-1073.
13. Renzoni D, Pugh D, Siligardi G, Das P, Morton C, Rossi C, Waterfield M, Campbell I, Ladbury J: Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase. Biochemistry 1996, 35(49):15646-15653.
14. Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R: Crystal structures of a complexed and peptide-free membrane protein–binding domain: molecular basis of peptide recognition by PDZ. Cell 1996, 85(7):1067-1076.
15. Hammarström A, Berndt KD, Sillard R, Adermann K, Otting G: Solution structure of a naturally-occurring zinc-peptide complex demonstrates that th N-terminal zinc-binding module of the Lasp-1 LIM domain is an independent folding unit. Biochemistry 1996, 35(39):12723-12732.
16. Glover JM, Harrison SC: Crystal structure of the heterodimeric bZIP transcription factor c-Fos–c-Jun bound to DNA. 1995.
17. Ghosh G, Van Duyne G, Ghosh S, Sigler PB: Structure of NF-κB p50 homodimer bound to a κB site. 1995.
18. Schwabe JW, Chapman L, Finch JT, Rhodes D: The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell 1993, 75(3):567-578.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT editor-in-chief@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE









