ABOUT AUTHORS:
Ketan M. Parmar*, Ritesh N. Sharma
S.K.Patel College of Pharmaceutical Education & research,
Department of Pharmaceutical chemistry, GANPAT UNIVERSITY.
*brave_student_90@yahoo.com
ABSTRACT
With the development of technologies to look at the expression levels of hundreds of miRNAs at a time and the clear role of miRNAs in cancers, groups began looking at miRNAs profiles of different cancers,especially the circulating miRNAs. We intended to make sure whether circulating miRNAs could be a promising biomarker of human cancers. Method: We comprehensively searched the Cochrane Library, Medline and EMbase from 1966 to Nov 2009 for the following terms: (“miRNA” or “microRNA”) and (“tumor” or “carcinoma”) and (“plasma” or “serum” or “circulating”). Detailed information was extracted from studies that met the inclusion criteria: blood-based miRNAs in human cancers and studies published in the English literature. Results: The current review show that different researches use different measurement methods which might impact the results;Cancers treatment might have an effect on circulating miRNAs; some miRNAs are multi-faceted RNA; small sample size might produce selection bias. Furthermore, because of the lack of randomized controlled trials and the heterogeneous nature of the available data, no attempt was made to perform quantitativemeta-analyses.
In this review, based on those researches, circulating miRNAs are promising and difficulties for their future application for diagnosing human cancers.
REFERENCE ID: PHARMATUTOR-ART-1739
INTRODUCTION
microRNA(abbreviated miRNA) is a short ribonucleic acid(RNA) molecule found in eukaryotic cells. A microRNA molecule has very few nucleotides(an average of 22) compared with other RNAs.(1)
miRNAs are post-transcriptional regulators that bind to complementary sequences on target messenger RNA transcripts(mRNAs), usually resulting in translational repression or target degradation and gene silencing. The human genome may encode over 1000 miRNAs, which may target about 60% of mammalian genes and are abundant in many human cell types.(2)
miRNAs show very different characteristics between plants and metazoans. In plants, repressions on the transcriptional level usually require a perfect or near-perfect target match, while a mismatched target can lead to gene silencing at the translational level. In metazoans, on the other hand, miRNA complementarity typically encompasses the 5' bases 2-7 of the microRNA, the microRNA seed region, and one miRNA can target many different sites on the same mRNA or on many different mRNAs. Another difference is the location of target sites on mRNAs. In metazoans, the miRNA target sites are in the three prime untranslated regions(3'UTR) of the mRNA.(3)
The first miRNAs were characterized in the early 1990s. However, miRNAs were not recognized as a distinct class of biological regulators with conserved functions until the early 2000s. Since then, miRNA research has revealed multiple roles in negative regulation (transcript degradation and sequestering, translational suppression) and possible involvement in positive regulation (transcriptional and translational activation). By affecting gene regulation, miRNAs are likely to be involved in most biological processes. Different sets of expressed miRNAs are found in different cell types and tissues.(4)
Aberrant expression of miRNAs has been implicated in numerous disease states, and miRNA-based therapies are under investigation.(5)
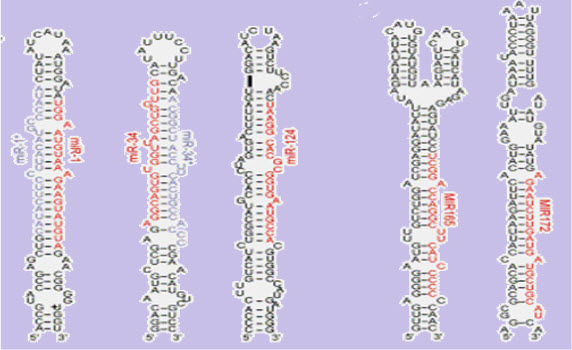
(FIGURE 1 - The stem-loopsecondary structureof a pre-microRNA from Brassica oleracea.(source - Lynan-Lennon BiolRev CambPhilosSoc. 2009 Feb;84(1):55-71)
History(6)
MicroRNAs were discovered in 1993 by Victor Ambros, Rosalind Lee and Rhonda Feinbaum during a study of the gene lin-14 in C. elegans development. They found that LIN-14 protein abundance was regulated by a short RNA product encoded by the lin-4 gene. A 61-nucleotide precursor from the lin-4 gene matured to a 22-nucleotide RNA that contained sequences partially complementary to multiple sequences in the 3’ UTR of the lin-14 mRNA. This complementarity was both necessary and sufficient to inhibit the translation of the lin-14 mRNA into the LIN-14 protein. Retrospectively, the lin-4 small RNA was the first microRNA to be identified, though at the time, it was thought to be a nematode idiosyncrasy. Only in 2000 was a second RNA characterized: let-7, which repressed lin-41, lin-14, lin-28, lin-42, and daf-12 expression during developmental stage transitions in C. elegans. let-7 was soon found to be conserved in many species, indicating the existence of a wider phenomenon.
Nomenclature(7)
Under a standard nomenclature system, names are assigned to experimentally confirmed miRNAs before publication of their discovery. The prefix "mir" is followed by a dash and a number, the latter often indicating order of naming. For example, mir-123 was named and likely discovered prior to mir-456. The uncapitalized "mir-" refers to the pre-miRNA, while a capitalized "miR-" refers to the mature form. miRNAs with nearly identical sequences except for one or two nucleotides are annotated with an additional lower case letter. For example, miR-123a would be closely related to miR-123b. Pre-miRNAs that lead to 100% identical mature miRNAs but that are located at different places in the genome are indicated with an additional dash-number suffix. For example, the pre-miRNAs hsa-mir-194-1 and hsa-mir-194-2 lead to an identical mature miRNA099(hsa-miR-194) but are located in different regions of the genome. Species of origin is designated with a three-letter prefix, e.g., hsa-miR-123 is a human (Homo sapiens) miRNA and oar-miR-123 is a sheep (Ovisaries) miRNA. Other common prefixes include 'v' for viral (miRNA encoded by a viral genome) and 'd' for DrosophilamiRNA (a fruit fly commonly studied in genetic research).
When two mature microRNAs originate from opposite arms of the same pre-miRNA, they are denoted with a -3p or -5p suffix. (In the past, this distinction was also made with 's' (sense) and 'as' (antisense)). When relative expression levels are known, an asterisk following the name indicates an miRNA expressed at low levels relative to the miRNA in the opposite arm of a hairpin. For example, miR-123 and miR-123 would share a pre-miRNA hairpin, but more miR-123 would be found in the cell.
Mechanisms of miRNA-mediated repression
The mature strand of the miRNA is incorporated into a complex of ribonucleotide protein to form the miRNP , also called the miRNA – induced silencing complex (miRISC). The primary protein in this complex are members of the argonate (AGO)family,each of which possesses repressive capabilities.Mammals have four AGO protein (AGO1 – AGO4)of which only AGO2 has the potential to cleva target sequences due to its RNaseH-like domain (Peters and Meister 2007).
The mature miRNA is used as a guide in the miRNP to recognize its target mRNA, to which it may be complementary with different degrees. In plants , miRNAs exibit a near – perfest match to target,thereby triggering an RNAi – like mechanism that result in cleavage of target mRNAs.
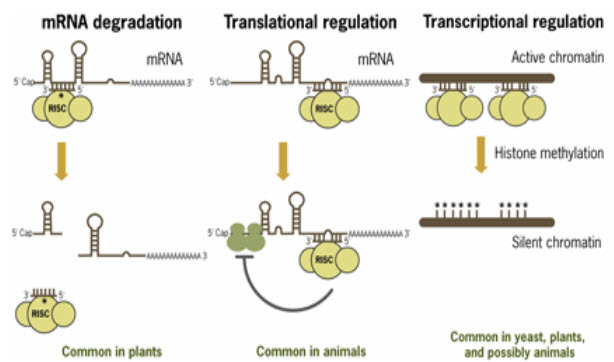
(FIGURE 2 - Mechanisms of miRNA-mediated post-transcriptional regulation (Source –www.ambion.com)
Biogenesises(8)
The majority of the characterized miRNA genes are intergenic or oriented antisense to neighboring genes and are therefore suspected to be transcribed as independent units.However, in some cases a microRNA gene is transcribed together with its host gene; this provides a mean for coupled regulation of miRNA and protein-coding gene.As much as 40% of miRNA genes may lie in the introns of protein and non-protein coding genes or even in exons of long nonprotein-coding transcripts. These are usually, though not exclusively, found in a sense orientation, and thus usually are regulated together with their host genes .Other miRNA genes showing a common promoter include the 42-48% of all miRNAs originating from polycistronic units containing multiple discrete loops from which mature miRNAs are processed, although this does not necessarily mean the mature miRNAs of a family will be homologous in structure and function.
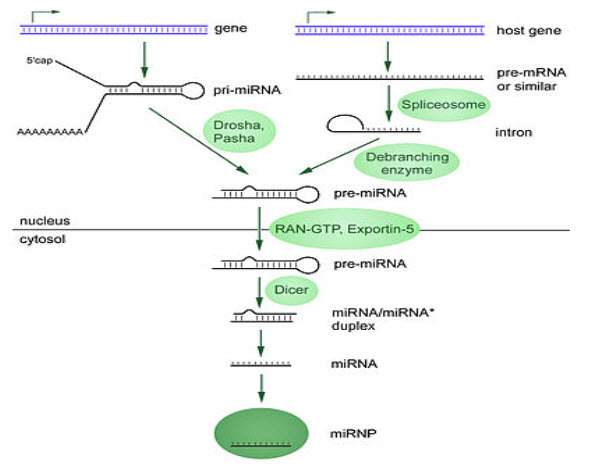
(FIGURE 3 – Biogenesis of miRNAs ( Source - Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (January 2006)
MicroRNAs are produced from either their own genes or from introns.
The promoters mentioned have been shown to have some similarities in their motifs to promoters of other genes transcribed by RNA polymerase II such as protein coding genes. The DNA template is not the final word on mature miRNA production: 6% of human miRNAs show RNA editing (IsomiRs), the site-specific modification of RNA sequences to yield products different from those encoded by their DNA. This increases the diversity and scope of miRNA action beyond that implicated from the genome alone.
Transcription(9)
miRNA genes are usually transcribed by RNA polymerase II(Pol II). The polymerase often binds to a promoter found near the DNA sequence encoding what will become the hairpin loop of the pre-miRNA. The resulting transcript is capped with a specially-modified nucleotide at the 5’ end, polyadenylated with multiple adenosines (a poly(A) tail), and spliced. Animal miRNAs are initially transcribed as part of one arm of an ∼80 nucleotide RNA stem-loop that in turn forms part of a several hundred nucleotides long miRNA precursor termed a primary miRNA (pri-miRNA)s. When a stem-loop precursor is found in the 3’ UTR, a transcript may serve as a pri-miRNA and a mRNARNA polymerase III(Pol III) transcribes some miRNAs, especially those with upstream Alu sequences, transfer RNAs(tRNAs), and mammalian wide interspersed repeat (MWIR) promoter units.
Nuclear processing(10)
A single pri-miRNA may contain from one to six miRNA precursors. These hairpin loop structures are composed of about 70 nucleotides each. Each hairpin is flanked by sequences necessary for efficient processing. The double-stranded RNA structure of the hairpins in a pri-miRNA is recognized by a nuclear protein known as DiGeorge Syndrome Critical Region 8(DGCR8 or "Pasha" in invertebrates), named for its association with DiGeorge Syndrome. DGCR8 associates with the enzyme Drosha, a protein that cuts RNA, to form the "Microprocessor" complex. In this complex, DGCR8 orients the catalytic RNase III domain of Drosha to liberate hairpins from pri-miRNAs by cleaving RNA about eleven nucleotides from the hairpin base (two helical RNA turns into the stem). The product resulting has a two-nucleotide overhang at its 3’ end; it has 3' hydroxyl and 5' phosphate groups. It is often termed as a pre-miRNA (precursor-miRNA).
pre-miRNAs that are spliced directly out of introns, bypassing the Microprocessor complex, are known as "Mirtrons." Originally thought to exist only in Drosophila and C. elegans, mirtrons have now been found in mammals.
Nuclear export(11)
pre-miRNA hairpins are exported from the nucleus in a process involving the nucleocytoplasmic shuttle Exportin-5. This protein, a member of the karyopherin family, recognizes a two-nucleotide overhang left by the RNase III enzyme Drosha at the 3' end of the pre-miRNA hairpin. Exportin-5-mediated transport to the cytoplasm is energy-dependent, using GTP bound to the Ran protein.
Cytoplasmic processing(12)
In cytoplasm, the pre-miRNA hairpin is cleaved by the RNase III enzyme Dicer. This endoribonuclease interacts with the 3' end of the hairpin and cuts away the loop joining the 3' and 5' arms, yielding an imperfect miRNAmiRNAduplex about 22 nucleotides in length. Overall hairpin length and loop size influence the efficiency of Dicer processing, and the imperfect nature of the miRNAmiRNA pairing also affects cleavage. Although either strand of the duplex may potentially act as a functional miRNA, only one strand is usually incorporated into the RNA-induced silencing complex (RISC) where the miRNA and its mRNA target interact.
The RNA-induced silencing complex
The mature miRNA is part of an active RNA-induced silencing complex (RISC) containing Dicer and many associated proteins. RISC is also known as a microRNA ribonucleoprotein complex (miRNP); RISC with incorporated miRNA is sometimes referred to as "miRISC."
Dicer processing of the pre-miRNA is thought to be coupled with unwinding of the duplex. Generally, only one strand is incorporated into the miRISC, selected on the basis of its thermodynamic instability and weaker base-pairing relative to the other strand. The position of the stem-loop may also influence strand choice. The other strand, called the passenger strand due to its lower levels in the steady state. In some cases, both strands of the duplex are viable and become functional miRNA that target different mRNA populations.(13)
Members of the Argonaute(Ago) protein family are central to RISC function. Argonautes are needed for miRNA-induced silencing and contain two conserved RNA binding domains: a PAZ domain that can bind the single stranded 3’ end of the mature miRNA and a PIWI domain that structurally resembles ribonuclease-H and functions to interact with the 5’ end of the guide strand.
They bind the mature miRNA and orient it for interaction with a target mRNA. Some argonautes, for example human Ago2, cleave target transcripts directly; argonautes may also recruit additional proteins to achieve translational repression. The human genome encodes eight argonaute proteins divided by sequence similarities into two families: AGO (with four members present in all mammalian cells and called E1F2C/hAgo in humans), and PIWI (found in the germ line and hematopoietic stem cells).
Additional RISC components include TRBP[human immunodeficiency virus (HIV) transactivating response RNA (TAR) binding protein, PACT (protein activator of the interferon induced protein kinase (PACT), the SMN complex, fragile X mental retardation protein (FMRP), Tudor staphylococcal nuclease-domain-containing protein (Tudor-SN), the putative DNA helicase MOV10, and the RNA recognition motif containing protein
Potential miRNAs Functions and targets
|
MiRNA |
Dysregulation |
Function |
Validate targets |
Oncogen or Tumour suppression |
|
miR – 154 , miR – 16 – 1 |
Loss in CLL,Prostate cancer & multiple myolema |
Induces apoptosis , Inhibit tumorigenesis. |
BCL 2, WTI,RAB9B & MAGE 83 |
TS |
|
Let – 7 (a,b,c,d,e,f,g&i) |
Loss in lung &brest cancer & in various solid & haematopoietic malignancies |
Induces apoptosis , Inhibit tumorigenesis. |
RAS , MTC & HMGA 2 |
TS |
|
miR - 29 (a,b&c) |
Loss in aggressive CLL , breast cancer |
Induces apoptosis , Inhibit tumorigenesis |
TCLI , MCLI & DNMTS |
TS |
|
miR – 34 |
Loss in pancreatic , colon , Liver cancer |
Induces apoptosis |
CDK4 , CDK6, CYCLIN E2 & MET |
TS |
|
miR – 145 |
Loss in breast cancer |
Induces apoptosis , Inhibit proliferation |
ERG |
TS |
|
miR – 221 , miR– 122 |
Loss in erythroblasticleukemia |
Inhibit proliferation in erythroblast |
KIT |
TS |
|
miR– 21 |
Upregulatated in glioblastomas , aggressive CLL, breast, colon , prostste& stomach cancer. |
Inhibit apoptosis & increase tumorigenesis |
PPEN , PDCD 4, TPMI & TIMP 3 |
ONC |
|
miR – 372 &miR– 373 |
Upregulated in Testicular tumors |
Promotes tumorigenicity in cooperation with ras |
LATS 2 |
ONC |
miRNA AND DISEASE(14)
Just as miRNA is involved in the normal functioning of eukaryotic cells, so has dysregulation of miRNA been associated with disease. A manually curated, publicly available database, miR2Disease, documents known relationships between miRNAdysregulation and human disease.
miRNA and inherited diseases(15)
A mutation in the seed region of miR-96 causes hereditary progressive hearing loss. A mutation in the seed region of miR-184 causes hereditary keratoconus with anterior polar cataract.Deletion of the miR-17~92 cluster causes skeletal and growth defects.
miRNA and heart disease(16)
The global role of miRNA function in the heart has been addressed by conditionally inhibiting miRNA maturation in the murine heart, and has revealed that miRNAs play an essential role during its development. miRNA expression profiling studies demonstrate that expression levels of specific miRNAs change in diseased human hearts, pointing to their involvement in cardiomyopathies. Furthermore, studies on specific miRNAs in animal models have identified distinct roles for miRNAs both during heart development and under pathological conditions, including the regulation of key factors important for cardiogenesis, the hypertrophic growth response, and cardiac conductance.
miRNA and the nervous system(17)
miRNAs appear to regulate the nervous system. Neural miRNAs are involved at various stages of synaptic development, including dendritogenesis (involving miR-132, miR-134 and miR-124), synapse formation and synapse maturation (where miR-134 and miR-138 are thought to be involved). Some studies find altered miRNA expression in schizophrenia.
miRNA and cancer(18)
Several miRNAs have been found to have links with some types of cancer and are sometimes referred to as "oncomirs." MicroRNA-21 was one of the first microRNAs to be identified as an oncomir.
A study of mice altered to produce excess c-Myc— a protein with mutated forms implicated in several cancers — shows that miRNA has an effect on the development of cancer. Mice that were engineered to produce a surplus of types of miRNA found in lymphoma cells developed the disease within 50 days and died two weeks later. In contrast, mice without the surplus miRNA lived over 100 days. Leukemia can be caused by the insertion of a viral genome next to the 17-92 array of microRNAs leading to increased expression of this microRNA.
Another role for miRNA in cancers is to use their expression level as a prognostic, for example one study on NSCLC samples found that low miR-324a levels could serve as a prognostic indicator of poor survival, another found that either high miR-185 or low miR-133b levels correlated with metastasis and poor survival in colorectal cancer.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
Circulating miRNAs in Human Cancers
It is reported that miRNAs could be an ideal class of blood-based biomarkers for cancerdetection because: (i)miRNA expression is frequently dysregulated in cancer (ii) expression patterns of miRNAs in human cancer appear tobe tissue-specific and (iii) miRNAs have unusually high stability in formalin-fixed tissues This third point led us to speculatethat miRNAs may have exceptional stability in plasma and be promising biomarkers for diagnosing human cancers.
Leukemia(19)
MiRNAs play a very important role in normal hematopoiesis because they regulate hematopoietic differentiation in almost every stage, while their aberrant expression has been associated with many diseases, including hematological malignancies. Increasing evidence has shown that miRNAs could function as either tumor suppressors or oncogenes in cancers such as leukemia, while other miRNAs might be beneficial for diagnosis and prognosis, predicted to be newly developed biomarkers.
Acute lymphoblastic leukemia(20)
A malignant disorder of lymphoid progenitor cells, affects both children and adults,with peak prevalence between the ages of 2 and 5 years. The precise pathogenetic events leading to development of acute lymphoblastic leukaemia are unknown Therefore, it is hard to diagnose leukaemia in the early stage. Recently, it has been reported that miRNAs are circulating in serum and tumor-derived miRNAs such as miR-155,miR-21, miR-15b, miR-16 and miR-24 are detected in the plasmas and serums of tumorpatients.These might be a new class of effective biomarkers, and we expect that the miRNAs abundance profile in plasma might reflect physiological and/or pathological conditions.
Chronic lymphocyticleukemia(21)
This accounts for around 30% of leukaemia cases in the United States of America, with the highest incidence in the elderly population. The age-adjusted incidence rate of chronic lymphocytic leukaemia is 4.0 per 100,000 men and women per year. Clinically, Chronic lymphocytic leukemia is a heterogeneous disease. Consistent with clinical heterogeneity, a number Of molecular prognostic factors have been defined for B cell chronic lymphocytic leukemia, most notably mutation status of the IGHV locus, ZAP-70 expression and recurrent cytogenetic lesions.
Hodgkin lymphoma(22)
There are a few studies referring to miRNAs alterations in Hodgkin Lymphoma. analyzed Hodgkin lymphoma cell lines and tissue samples and detected high levels of BIC and mir-155 in Hodgkin lymphoma. Thereafter, the first description of circulating miRNAs that had potential as non-invasive diagnostic markers in Hodgkin lymphoma for was made in 2008 when Lawrie et al investigated tumor-associatedmiR-155, miR-210 and miR-21 in serum of diffuse large B-cell lymphoma (Hodgkin lymphoma) patients and healthy controls.
Lung cancer(23)
With more than 215,000 new cancer cases and more than 160,000 cancer deaths estimated in 2008, lung carcinoma continues to be the leading cause of cancer mortality in the United States. Despite potentially curative surgery, about 40% of patients will relapse within 5 years . Cancers, including lung cancer and colorectal cancer, are often diagnosed at a late stage with concomitant poor prognosis. Although tumor markers greatly improve diagnosis, the invasive, unpleasant, and inconvenient nature of currentdiagnostic procedures limits their application. Hence, there is a great need for identification of novel non-invasive biomarkers for early tumor detection.
A large number of researches have found that significant differences were presented between the exosomalmiRNA levels for the lung adenocarcinoma group and the control group found that miRNA expression profiles might be diagnostic and prognostic markers of lung cancer.
Ovarian cancer(24)
In 2008, it was expected that 20,180 women will be diagnosed with ovarian cancer and 15,310 will succumb to the disease . Ovarian cancer is a devastating illness in which only 20% of patients are diagnosed with stage I disease.
The poorprognosis associated with ovarian cancer is multi-factorial; a lack of minimally invasive, early detection tests, subtle symptom development and tumor chemo-resistance. Even with the advent of chemo-resistance assays it is still difficult to predict drug resistance and only 10–15% of patients will remain in prolonged remission after initial cytotoxic therapy.
While annual pelvic examination is widely practiced, it lacks the sensitivity to be used a screening strategy for ovarian cancer. Women at high risk for ovarian cancer may typically undergo screening with trans-vaginal ultrasound and serum CA-125. CA-125, however, remains a poor marker for early stage disease with a documented sensitivity of 40% .
Multiple recent profiling studies also indicate that miRNA expression is significantly changed in ovarian cancer.
Breast cancer(25)
Breast carcinoma, which is the second most prevalent cancer in women, is diagnosed in >200,000 woman in the USA every year. Early detection is a major factor contributing to the 2.3% annual decline in breast cancer death rates over the past 10 years Nonetheless 40,480 women in the USA were projected to die from breast cancer in 2008 in part because currently available breast cancer screening tools such as mammography and breast examination miss10-40% of early breast cancers and are least effective in detecting cancer in young women, whose tumors are often more aggressive.
An invasive needle or surgical biopsy must be performed when an area of suspicion is identifiedin order to confirm, by cytologic or histologic evaluation, the presence of malignancy, even though 66–85% of abnormalities are benign. To date, only two markers have been established so far in the routine assessment of breast cancer: ER Although these markers are currently available, ER and HER2 assessment is far from perfect A number of circulating tumour markers and carbohydrate antigen 15-3 are used clinically in the management of breast cancer, but the sensitivity of these markers is low, so that they are not useful as screening tools though they have long been in clinical use as prognostic markers and to monitor for disease progression or recurrence.Alterations in miRNA expression have been associated with tumor suppression or tumorigenesis, metastasis and poor prognosis in human breast cancer.
Other cancers(26)
Furthermore, in patients with squamous cell carcinoma of the tongue, plasma levels of miR-184 were significantly higher than those in healthy individuals, and miR-184 levels were significantly reduced after surgical removal of the primary tumors .Moreover, miR21, known to be over expressed in gliobalstomatumors was elevated in serum microvesicles from glioblastoma patients. There is no related clinical trialin melanoma, pancreatic cancer,
SUMMARY
miRNAs control cell cycle, cell differentiation and apoptosis by regulating oncogenes and tumor supressor genes. miRNAs are misexpressed in cancer and are therefore excellent diagnostic/ prognostic markers in cancer. Some miRNAs e.g. mir-155, can cause cancer and oncogenic miRNAs may be therapeutic targets in cancer .Other miRNAs like let-7, may prevent cancer and may be therapeutic molecules themselves.
REFRENCES
1.Bartel, D. P. (2009). "MicroRNAs: target recognition and regulatory functions". Cell136 (2): 215–233.
2.Kusenda, B.; Mraz, M.; Mayer, J.; Pospisilova, S. (2006). "MicroRNA biogenesis, functionality and cancer relevance"..
3.Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ (2009). "MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis"..
4.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (February 2006). "Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs". Nature433 (7027): 769–73. Bibcode2005Natur.433..769L.
5.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM (April 2009). "bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila".
6.Cuellar TL, McManus MT (December 2008). "MicroRNAs and endocrine biology". J. Endocrinol.187 (3): 327–32.
7.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (January 2006). "miRBase: microRNA sequences, targets and gene nomenclature". Nucleic Acids Res.34 (Database issue):
8.Faller, M.; Guo, F. (2008). "MicroRNA biogenesis: there's more than one way to skin a cat.". Biochimica et BiophysicaActa1779 (11): 663–667.
9.Winter, J.; Jung, S.; Keller, S.; Gregory, R.; Diederichs, S. (2009). "Many roads to maturity: microRNA biogenesis pathways and their regulation". Nature Cell Biology11 (3): 228–234.
10.Ohman, M. (2007). "A-to-I editing challenger or ally to the microRNA process". Biochimie89 (10): 1171–1176.
11.Murchison, E.; Hannon, G. (2007). "MiRNAs on the move: miRNA biogenesis and the RNAi machinery". Current opinion in cell biology16 (3): 223–229.
12.Lund, E.; Dahlberg, J. (2006). "Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs". Cold Spring Harbor symposia on quantitative biology71: 59–66.
13.Artmann, S.; Jung, K.; Bleckmann, A.; Beißbarth, T. (2012). Provero, Paolo. ed. "Detection of Simultaneous Group Effects in MicroRNA Expression and Related Target Gene Sets"..
14.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. (January 2009). "miR2Disease: a manually curated database for microRNA deregulation in human disease.". Nucleic Acids Research. 37 (Database issue) (Database issue): D98–104.
15.Mencía, Á.; Modamio-Høybjør, S.; Redshaw, N.; Morín, M. A.; Mayo-Merino, F.; Olavarrieta, L.; Aguirre, L. A.; Del Castillo, I. et al. (2009). "Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss". Nature Genetics41 (5): 609–613.
16.Akçakaya P, Ekelund S, Kolosenko I, et al. (August 2011). "miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer". Int J Oncol39 (2): 311–8.
17.Pheasant M, Mattick JS (September 2007). "Raising the estimate of functional human sequences". Genome Res.17 (9): 1245–53.
18.Raponi M, Dossey L, Jatkoe T, et al(2009). MicroRNA classifiers for predictingPrognosis of squamous cell lung cancer Cancer Res, 69, 5776-5783.
19.Reid BJ, Haggitt RC, Roessler S, Budhu A, Wang XW(2007). Future of molecular Profiling of huma hepatocellular carcinoma. Future Oncol, 3, 429-439.
20.Roulston JE(1990). Limitations of tumour markers in screening. Br Jsurg, 77,961-962.
21.Rubin CE, et al(1988). Observer variation in the diagnosis of dysplasia in Barrett’s esophagus. Hum Pathol, 19, 166-178.
22.Schetter AJ, Leung SY, Sohn JJ, et al(2008). MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma.
23.Sempere LF, Christensen M, Silahtaroglu A, et al(2007). Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer.
24.Tanaka M, Oikawa K, Takanashi M, et al(2009). Downregulation of miR-92 in human plasma is a novel marker for acute leukemia patients.
25.Thompson A, Brennan K, Cox A, et al(2008). Evaluation of the current knowledge limitations in breast cancer research: a gap analysis. Breast Cancer Res, 10, R26.
26.Yang N, Kaur S, Volinia S, et al(2008). MicroRNA microarrayidentifies Let-7i as anovel biomarker and therapeutic target in human epithelial ovarian cancer.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE









