ABOUT AUTHORS
Tahseen Sameena*1, Prathima Patil1, S.P.Sethy*1
1Department Of Pharmaceutics.
Azad College of Pharmacy
Moinabad-Chilkur Road , Hyderabad- India
* tahseensameena1992@gmail.com
{ DOWNLOAD AS PDF }
ABSTRACT: -
The Human Genome Project (HGP) refers to the international 13‐year effort, formally begun in October 1990 and completed in 2003, to discover all the estimated 20,000–25,000 human genes and make them accessible for further biological study. Another goal of this project was to determine the complete sequence of the 3 billion DNA subunits (bases in the human genome). As part of the HGP, parallel studies were carried out on selected model organisms such as the bacterium E.coli and the mouse to help develop the technology and interpret human gene function. The DOE Human Genome Program and the U.S National institute of Health (NIH) National Human Genome Research Institute (NHGRI) together sponsored the U.S.Human Genome Project.”
Reference Id: PHARMATUTOR-ART-2532
INTRODUCTION:
The most important feature of DNA molecule are the nucleotide sequences, and the identification of genes and their activity .Science 1920 scientist have been working to determine the sequences of a piece of DNA. This was further extended for complete sequence determination of genome of certain lower organism e.g. Plasmid pbr322. The sequencing of the human genome represented the largest single undertaking in the history of biological science and stands as a signature scientific achievement. All of history in the making, human DNA took just 13 years (1990-2003) [1] to sequence under the Human Genome Project (HGP), an international public project led by the United States, and a complementary private program. Sequencing the human Genome determining the complete sequence of the 3 billion DNA base pairs and identifying each human gene—required advanced technology development and the assembly of an interdisciplinary team of biologists, physicists, chemists, computer scientists, mathematicians and engineers. Scientists are using the reference genome, the knowledge of genome structure, and the data resulting from the HGP as the foundation for fundamental advancements in medicine and science with the goals of preventing, diagnosing, and treating human disease. Also, while foundational to the understanding of human biological systems, the knowledge and advancements embodied in the human genome sequencing, and the sequencing of model organisms, are useful beyond human biomedical sciences .The resulting “genomic revolution” is influencing renewable energy development, industrial biotechnology, agricultural biosciences, veterinary sciences, environmental science, forensic science and homeland security, and advanced studies in zoology, ecology, anthropology and other disciplines[2].
THE BIRTH AND ACTIVITY OF HUMAN GENOME PROJECT (HGP): The human genome project was conceived in 1984, and officially started in October 1990. The primary objective of human genome project was to determine the nucleotide sequence of entire human nuclear genome [3]. In addition to this HGP was also entrusted to elucidate the genome of several other model organisms like E.coli, S. Cerivisiae, Mus musculus (mouse). James Watson who elucidated the DNA structure was the first director of HGP. In 1997 US established the National Human Genome Research Institute (NHGRI). The HGP was an international venture involving research group of six countries- USA, UK, FRANCE, GERMANY, CHINA, and JAPAN and several individual research laboratory and scientists and technicians from various disciplines throughout the world. This collaborative venture was named as International Human Genome Sequencing Consortium headed by Francis Collins [4]. A total expenditure of 3 billion USD and a time period of 10-13 years of completion of HGP. A second human genome project was constituted by Celera Genomics, Maryland USA in 1998.The team was led by Craig Venter and a very rapid and un expected progress occurred in HGP with good co-operation between the team of workers and improved method of sequencing.
ANNOUNCEMENT OF THE DRAFT SEQUENCE OF HUMAN GENOME: The date 26th June 2000 will be remember as one of the most important dates in the history of science or even mankind. It was on this day Francis Collins and Craig Venter, the leader of two human genome projects in the presence of the president of US, jointly announced the working draft of human genome sequence. The detailed result was later published in scientific journal the nature and science. The human genome project results in worldwide attention. The achievement was hailed with many descriptions in media
• The mystery of life unraveled
• The library of life
• The periodic table of life
• The holy grail of human life
It may however, be noted that the draft human genome sequence was not complete and may represent around 90%. The remaining 10% is made up of sequence where few genes are
located [5].
APROACHES FOR GENOME SEQUENCING:
For elucidating the human genome different approaches were used by the two HGP group. IHGSC predominantly employed the map first and sequence later approach. The principal method was Hierarchical Short gun Method [6]. This method involves fragmentation of genome to small fragment (100-200Kb) and inserting them into vector e.g. BACs and cloning. Celera genomics used Whole Genome Shotgun Method. This bypasses the mapping step and saves time .Celera groups was lucky to have high-throughput put sequesters and powerful computer programmes that help for early completion of human genome sequence.
One of the most difficult questions of human genome project was whose is being sequenced and how it will relate to 6 billion people of the world with wide range of variation? There is no such simple answer to this question but looking from the positive side it does not matter whose genome was sequenced, since phenotypic difference between individuals are variations in just 0.1% of the total genome sequenced. Therefore many individual genomes can be used as a source material for sequencing. Much of the human genome work was performed on the material supplied by Centre of human Polymorphism in Paris, France. This institute had collected cell lines from sixty different French families each spanning three generations.
HUMAN GENOME SEQUNCE-RESULTS SUMMARISED:
The results of human genome project [7] is too vast and only some highlights we are presenting here Table-1
A list of principal method used for mapping of genome (For normal and disease gene)
|
METHOD |
COMMENTS |
|
DNA sequencing |
Physical map of DNA can be identified with high resolution |
|
Use of Probes |
To identify RFLPs,STS,and SNPs |
|
Radiation hybrid mapping |
Fragment genome into large pieces and locate markers |
|
Florescent in situ hybridization (FISH) |
To locate a gene on chromosome |
|
Sequence target site mapping |
Applicable to any part of DNA sequence if some sequence information is available |
|
Express sequence tag mapping |
A variant of STS mapping , expressed genes are actually mapped and located |
|
Pulsed-field electrophoresis(PFGE) |
For separation and isolation of large DNA fragment |
|
Cloning vectors(Plasmid,cosmid,phase,YACs BACs) |
to isolate DNA fragment of variable length |
|
Polymerase chain reaction |
To amplify gene fragment |
|
Chromosome walking |
Useful for cloning of overlapping DNA fragment |
|
Chromosome jumping |
DNA can be cut into larger fragment and circularized for use in chromosome walking |
|
Detection of cytogenetic abnormalities |
Certain genetic disease can be identified by cloning the affected genes |
|
Databases |
Existing database facilitates gene identification by comparison of DNA and protein sequence |
Table-2
Major highlights of human genome.
|
1. The draft represents about 90% of the entire human genome. It is believed that most of the entire parts have been identified. |
|
2. The remaining 10% of the genome sequences are the ends of chromosome (telomeres). |
|
3. Human genome is composed of 3200MB i.e. 3.2 billions base pairs |
|
4. Approx. 1.1 to 1.5% of the genome codes for proteins |
|
5. Approx. 24% of the total genome is composed of introns that split the coding regions (exons) and appear as a repeating unit with no specific functions[8]. |
|
6.The no of protein coding genes is in the range of 30,000-40,000[9] |
|
7. An average gene consists of 3000 base pair. Dystrophin gene is the largest known human gene containing 2.4million base. |
|
8. Chromosome 1 (The largest chromosome in humans) contains the highest number of genes (2968) while the Y chromosome has the lowest |
|
9.Gene and DNA sequences are associated with many disease such as breast cancer muscle disease deafness and blindness have been identified |
|
10. Repeated sequences constitute about 50% of human genome. |
|
11. A vast majority of human genome (97%) have unknown functions |
|
12. Between the humans , the DNA differ only by 0.2% or one in 500 base |
|
13. more than 3 million single nucleotide polymorphism have been identified |
|
14.Human DNA is about 98% identical with chimpanzees |
|
15. About 200 genes are close to that found in bacteria |
Table-3 Some interesting analogs/sidelight about human genome.
|
1. The base sequence in human genome would fill about 200 telephone book of 100 pages each. |
|
2. If the genome is recited at a rate of one base pair per second for 24 hours a day it would take a century to recite the book of life. |
|
3. If a typist types at a rate of 60 words per minute i.e. 360 letter for 8 hrs a day it would take 50 years to type the human genome. |
|
4. If the DNA sequence is typed in lines 10 cm containing 60 nucleotide base and printed the human genome sequence (from a single cell) would stretch at a distance of 5000 km. |
|
5. If the DNA in the entire human body is put end to end it would reach to the sun and back over 600 times. |
|
6. The total expenditure of human genome project was 3 billion USD. The magnitude of the huge amount has to be appreciated .If someone start counting at a nonstop rate of a dollar per second it would take about 90 years to complete. |
Table-4 Different categories of protein encoded by human genes (Based on human genome project report 2001) [10]
|
CATEGORY OF PROTEIN |
PERCENTAGE |
ACTUAL NO OF GENES |
|
Unknown functions |
41.0 |
12,809 |
|
Nucleic acid enzymes |
7.5 |
2,308 |
|
Transcription factor |
6.0 |
1,850 |
|
Receptors |
5.0 |
1,543 |
|
Hydrolases |
4.0 |
1,227 |
|
Regulatory protein |
3.2 |
988 |
|
Proto oncogenes |
2.9 |
902 |
|
Structural protein of cytoskeletons |
2.8 |
876 |
|
Kinase |
2.8 |
868 |
It is surprisingly to note that the number of genes found in humans is only twice that of round worm (19,099) and thrice that of fruit fly (13,001). Around 200 genes appear to have been derived from bacteria by lateral transfer. Surprisingly none of the genes are present in non-vertebrate eukaryotes. The protein encoded by human genes are more complex than that of invertibrates.The flood of data of human genome project will be highly useful for bio informatics and biotechnology.
Table-5 A selected list of genomes that have been sequenced [11]
|
NAME OF THE SPECIES |
GENOME SIZE(Mb/Kb) |
COMMENTS |
|
Bactriophase QX174 |
5.38Kb* |
First genome sequences1977 |
|
Plasmid Pbr322 |
4.3Kb |
First plasmid sequenced 1979 |
|
Yeast chromosome III |
315Kb |
First chromosome sequenced 1992 |
|
Haemophilus influenza |
1.8Mb* |
First genome of cellular organism to be sequenced 1995 |
|
Saccharomyces cerevisiae |
12Mb |
First eukaryotic organism to be sequenced1996 |
|
Arabidopsis thaliana |
125 Mb |
First plant genome to be sequenced 2000 |
|
Homo sapiens |
3200Mb |
First mammalian genome to be sequenced 2001 |
|
Oryza sativa (rice) |
430Mb |
First crop plant genome to be sequenced 2002 |
|
Mus musculis |
3300Mb |
Animal model closest to human 2003 |
Kb*- Kilo base pair Mb*- Mega base pair.
ECONOMIC IMPACT OF HUMAN GENOME SCIENCE[12] :
The human genome sequencing programs had direct expenditures totaling almost USD 3.8 billion (USD 5.6 billion in constant 2010 dollars). In addition, beyond the original HGP investments, both the National Institutes of Health (NIH) and the Department of Energy (DOE) made and continue to make significant investments in the science, instrumentation, and related applications stemming from the human genome work. In fact, in both current and constant 2010 dollars the federal investment in human genomics‐related or enhanced research over the last seven years (2004–2010) has actually exceeded the federal investment in the HGP by 28 percent. These additional expenditures also generated direct and indirect (multiplier effect) economic impacts in the U.S economy. The decoding of the human genome was both a technological as well as scientific achievement. The technologies that have empowered genome sequencing range from the gene sequencers themselves, to sample preparation technologies, sample amplification technologies, and a range of analytical tools and technologies. An industry has grown up to supply the scientific research community in the private sector, government and academia with the equipment, supplies and services required to conduct genomics research and development (R&D) and associated product development. This industry, of course, generates additional economic impacts. To evaluate these genomics‐enabled industry impacts in the U.S., Battelle constructed a “from the ground‐up” database of individual companies engaged within the sector. The employment of this industry base was used as the foundation for an input/output analysis to quantify the total impacts of these firms (in terms of direct and indirect output and employment and their multiplier effect). The results of the analysis show that—spurred by the original investment and technological development impetus of the human genome sequencing projects—a substantial economic sector has developed thereby benefiting the U.S. economy in terms of business volume, jobs, and personal income supporting American families. Battelle’s analysis of the impact of human genome sequencing includes separate assessments of the HGP impacts, post‐HGP expenditure impacts, and the impacts of the genomics‐enabled industry sector. These separate results are contained in the full report. Table-6 provides a summation of the combined impacts from these analyses for the 1988–2010 periods.
Table-6 Cumulative economic impact of human genome sequencing, 1988–2010 (in billions, 2010 USD) [13]
|
IMPACT |
EMPLOYMENT (job-years) |
PERSONAL INCOME |
OUTPUT |
STATE/ LOCAL TAX REVENUE |
FEDERAL TAX REVENUE |
|
Direct impact |
710,819 |
71.4 |
264.8 |
3.5 |
13.0 |
|
Indirect impact |
1,298,216 |
89.2 |
265.8 |
10.8 |
18.0 |
|
Induced impact |
1,818459 |
83.3 |
265.7 |
15.2 |
17.9 |
|
Total impact |
3,827495 |
243.9 |
796.3 |
29.5 |
48.9 |
|
Impact multiplier |
5.38 |
3.42 |
3.01 |
8.37 |
3.75 |
The direct impacts include nearly 711,000 direct job‐years, a combined personal income direct impact of more than USD 71 billion, and direct genomics‐driven output of nearly USD 265 billion. In Combined direct tax revenue more than USD 3.5 billion has been generated in state/local taxes and more than USD 13 billion in federal taxes. From just the perspective of federal revenue and expenses, the direct federal taxes generated to date (in 2010 USD) have exceeded the HGP and post‐HGP federal investments (USD 12.8 billion) by USD 166 million. The total employment impact exceeds 3.8 million job‐years over the 23‐year period examined. With an employment multiplier of 5.38, genomics research and associated industries generated an additional 4.38 jobs in the U.S. economy for every “genomics” job. This total level of impacted employment generated nearly USD 244 billion in personal income over the period in 2010 dollars amounting to an average of USD 63,700 in personal income per job‐year. The genome sequencing projects, associated research and industry activity generated a total economic (output) impact of more than USD 796 billion over the 1988–2010 periods. Considering the federal investment of USD 3.8 billion in the Human Genome Project from 1988–2003 (USD 5.6 billion in 2010 USD ) the HGP generated a return on investment (ROI) to the U.S. economy of 141 to 1-every USD 1 of HGP investment has helped to generate USD 141 in the economy.
Determining the full “current” economic impact of a program as complex and game‐changing as the sequencing of the human genome requires analysis of a time series of investments and organizational information extending back to the formal initiation of the Human Genome Project in 1990 and even slightly before. Table 7 shows the annual federal funding for the HGP . The combined annual federal investment in the HGP reached USD 3.8 billion in current dollars and when adjusted for inflation totals more than USD 5.6 billion in constant 2010 dollars.
Table 7 U.S. Human genome project federal funding (in millions USD)
|
Year |
DOE* (CURRENT USD) |
NIH* (CURRENT USD) |
U.S.FEDERAL (CURRENT USD) |
U.S.FEDERAL (CONSTANT USD) |
|
1988 |
10.7 |
17.2 |
29.7 |
54.0 |
|
1989 |
18.5 |
28.2 |
46.7 |
88.2 |
|
1990 |
27.2 |
59.5 |
86.7 |
160.0 |
|
1991 |
47.4 |
87.4 |
134.8 |
243.1 |
|
1992 |
59.4 |
104.8 |
164.2 |
289.5 |
|
1993 |
63.0 |
106.1 |
169.1 |
291.6 |
|
1994 |
63.3 |
127.0 |
190.3 |
321.2 |
|
1995 |
68.7 |
153.8 |
222.5 |
367.6 |
|
1996 |
73.9 |
169.3 |
243.2 |
390.5 |
|
1997 |
77.9 |
188.9 |
266.8 |
425.5 |
|
1998 |
85.5 |
218.3 |
303.8 |
448.5 |
|
1999 |
89.9 |
225.7 |
315.6 |
453.6 |
|
2000 |
88.9 |
271.7 |
360.6 |
490.1 |
|
2001 |
86.4 |
308.4 |
394.8 |
523.3 |
|
2002 |
90.1 |
346.7 |
434.3 |
548.4 |
|
2003 |
64.2 |
372.8 |
437.0 |
552.9 |
|
Total |
1,015.0 |
2,785.8 |
3,789.3 |
5,647.9 |
DOE*- Department Of Energy NIH*- National Institute of Health
Fig-1
The structure of forward and backward linkage impacts associated with the human genome sequencing
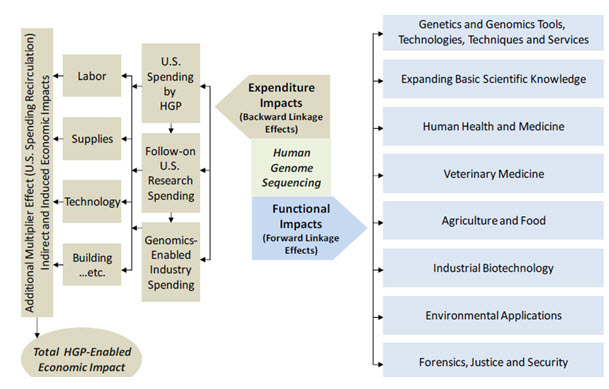
The economic impact analysis reported here in uses an I/O model to represent the inter relationships among economic sectors. I/O data show the flow of commodities to industries from producers and institutional consumers for any given region (in this case the United States as a whole). The data also model consumption activities by workers, owners of capital, and imports from outside the region. These trade flows built into the model permit estimating the impacts of one sector on other sectors. The impacts consist of three impact types: Direct (the specific expenditures impact of the program and/or sector(s) in question, Indirect (the impact on suppliers to the focus industry or program), and Induced (the additional economic impact of the spending of these suppliers and employees in the overall economy). The estimated impacts of sequencing the human genome were calculated using a U.S specific I/O model, the IMPLAN system, from MIG, Inc. IMPLAN (IMPLAN is one of the most widely used models in regional economics and can be used to analyze the economic impacts of companies, projects, or entire industries.) provides a software system and geographic specific data regarding economic sector interactions for calculating economic impacts. The model incorporates detail of more than 420 individual industry sectors that cover the entire national economy. With this coverage of 420 sectors, Battelle is able to model the cross‐sector economic activity that occurred throughout the economy as a result of the human genome sequencing work. Additionally, the IMPLAN model has built in economic “inflators” and “deflators” to allow for the development of cumulative multi-year impact estimation for the 23 years included in the analysis. Six key economic sectors were used in the development of the overall impact analysis as shown in Table 8. The unique nature of federal R&D investment, and the associated economic transactions and impact flowing from these investments, caused Battelle to select a single industry component “Scientific R&D services” as the most representative of these economic impacts.
Table 8 Sectors used in overall genomics impact assessment
|
GENOMIC SECTOR |
IMPLAN MODELING SECTOR |
|
Genomics- related bioinformatics |
Custom computer programming services |
|
Genomics and related testing |
Medical and diagnostic lab |
|
Genomic-related biologics and diagnostic substance |
Biologics and diagnostics |
|
Genomic instruments and equipment |
Analytical laboratory instrument manufacturing |
|
Genomics R&D/Genomics biology |
Scientific R&D services |
|
Drugs and pharmaceuticals |
Drugs and pharmaceuticals |
Historical R&D funding data includes federal HGP investments Table-7 and additional external and intramural federal investments in genomics areas after the 2003 end of the HGP by the NIH and DOE. To conservatively estimate additional genomics research investments by the major pharmaceutical manufacturers, an increasing share of the sector’s R&D funds was used over the 2004–2010 period. This was determined to be the most appropriate approximation of the pharmaceutical industry’ genomics involvement. While sales of genomics‐based drugs (or their related diagnostics) were considered, this would leave out significant past efforts and pipeline investments that have yet to yield sales. Historical employment data were developed using the unique properties of the National Establishment Time‐Series (NETS) database developed by Walls & Associates (built upon annual records from Dun & Bradstreet). Using a variety of current and historical sources and listings of genomics firms, a database of firms (where genomics‐specific activities, products, and services are the principal activity of the firm) was compiled. Using the NETS database a longitudinal set of employment metrics (1993–2010) were developed for each of the five“genomics” sectors listed in Table 9. The impact of genomics on the drugs and pharmaceuticals sector is estimated using only the R&D performance as described in the previous paragraph and not using employment. The NETS database allowed Battelle to capture the recent and historic employment of genomics firms, including those firms that were acquired or that have gone out of business over the last two decades. Another key component of the NETS database for this purpose is the “establishment” level information allowing for the inclusion of “genomic‐specific” operations of larger multi establishment firms to be included without having to include the firm’s total employment. This feature was particularly important in the Genomic & Related Testing sector. In the analysis that follows Battelle presents the direct effect values driving the model; addit induced). The following data are provided for each impact estimation: employment, personal income (including both wages and benefits), economic output, state and local tax revenue (including income and property taxes), and federal tax revenue (including contributions to Social Security). An impact multiplier is also provided for each type of data—for every one (job or dollar) of direct effect, the multiplier number will equal the total (including the direct effect) number of jobs or dollars created in the U.S. economy.
Table 9
Employment estimates of genomics sectors (key years)
|
“Genomics” sector |
2010 |
2003 |
2000 |
1993 |
|
Genomics R&D/Genomics Biotech |
13,323 |
13,140 |
8,275 |
2,378 |
|
Genomic Instruments & Equipment |
11,704 |
15,724 |
10,957 |
9,917 |
|
Genomic‐Related Biologics & Diagnostic Substances |
7,234 |
9,427 |
7,145 |
2,243 |
|
Genomic & Related Testing |
5,142 |
1,644 |
1,301 |
542 |
|
Genomics‐Related Bioinformatics |
797 |
1,430 |
667 |
174 |
|
Total Employment |
37,200 |
41,365 |
28,345 |
15,254 |
The NETS data records illuminate unique trends and changes apparent in the genomics sectors. A decline in bioinformatics employment between 2003 and 2010 occurs due to both declines in the overall information technology industry, but also through the incorporation of early bioinformatics companies into larger entities. Similarly, a decline in genomic instruments and equipment appears to be due to the continued merger and acquisition activity in this sector. Also apparent is the growth in genomics and related testing that has occurred since the completion of the HGP. Finally, it is likely that the recent recession has also continued to dampen employment levels across the board in 2010. Sector employment peaked in 2004 at 42,288. For the pharmaceutical industry, Table -10 projects the estimated genomics‐related R&D as well as total Pharmaceutical R&D for three key years.Given the magnitude of pharmaceutical R&D the Battelle team approached the use of these data with conservatism both in terms of the share of genomics R&D and the year in which these shares were first implemented in the model (2000).
Table 10 Pharmaceutical R&D estimates (key years, in millions current USD)
|
Pharmaceutical R&D |
2010 |
2003 |
2000 |
|
Genomics and related pharmaceutical R&D |
9,109 |
2,337 |
743 |
|
Total Pharmaceutical R&D |
45,547 |
28,853 |
24,754 |
FUNCTIONAL IMPACT OF HUMAN GENOME SCIENCE[14]:
The human sequencing programs sought to create benefits for humankind by illuminating the basic molecular processes governing life. It was expected that the resulting advancements in genomics knowledge and technologies would benefit human healthcare, energy, and multiple other fields. In this report we call these benefits functional impacts. Figure 2 illustrates the detailed structure of the functional impacts generated by the sequencing of the human genome. There was significant development of genomics tools, technologies and techniques to propel the sequencing efforts forward. These have, in many cases, been commercialized and form the foundation for a highly active and growing commercial genomics‐based industry. The application of these genomic tools, technologies and techniques resulted in a truly dramatic expansion of basic biological knowledge. As mentioned the full human genome sequence unveiled a complex biological system unanticipated by most in science, and has been a paradigm shifting event for biology. Some of the high profile impacts on basic sciences are highlighted in this chapter using illustrative examples and case studies.
Fig-2 The structure of functional impacts associated with the human genome sequencing [15]
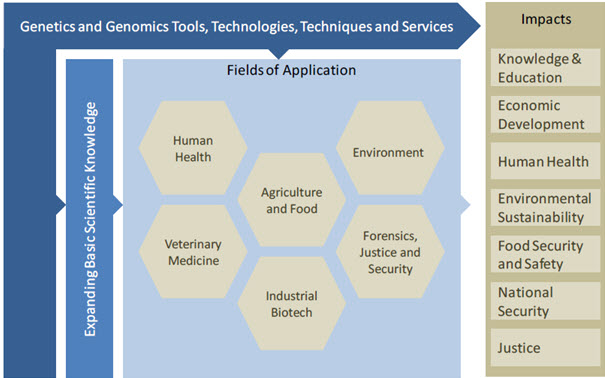
The sequencing of the human genome has resulted in a distinct paradigm shift in our understanding of the biology of humans, and indeed all organisms. As such, ipso factor the decoding of the human genome stands among the preeminent findings in the history of science.
Until the human genome sequence was published, and its implications understood, the reigning model of thinking in biological science was that biological forms are rendered under a very large but basically linear instruction set coded within DNA. It was expected that life forms and processes result from genes; that each gene generates an RNA template which specifies one protein, and these proteins (greatly simplified) assemble as “life.” The human genome sequence has overturned this one directional, simple view of the structure of life. It is now known that:
• The same gene can code for multiple proteins.
• Humans contain far less genes than originally anticipated (about 21,000 instead of upwards of 100,000 to 300,000 once anticipated), yet because of complex processes, such as alternative splicing and post‐translational modifications, the result may be over1 million different protein forms.
• Genes interact with one another and code for multiple RNAs.
• The structural genomic variations between organisms are relatively minor (the human and the chimpanzee, for example, differ from each other by only 1% of the DNA sequence), and thus the great variation found across organism structures is a result of regulatory processes.
• What had been termed “junk DNA” is typically not junk at all and plays important roles
In transcription and translational regulation of protein folding.
• Most common diseases, such as cancers, heart disease and psychiatric disorders are not homogeneous diseases, but differ dramatically across individual genomes from patient to patient.
GENOMICS DISCIPLINES:
The National center for Biotechnology Information at the National Library of Medicine notes that genomics itself has three primary branches, comprising:
• Structural Genomics – Includes mapping and sequencing
• Comparative Genomics – Including genetic diversity and evolutionary studies
• Functional Genomics – The study of the roles of genes in biological systems NEW AND EMERGING DISCIPLINES [16]:
• The ‘Omics – At the head of the list, is not one but a series of disciplines that are an out growth of genomics and its discoveries, generally grouped by the term ‘Omics. They Comprise: 1. Proteomics – The study of the set of proteins encoded by a genome 2. Metabolomics – The study of the complete collection of metabolites present in a cell Or tissue under a particular set of conditions
3, Transcriptomics-The study of all RNA molecules, including mRNA, rRNA, tRNA and non-coding RNA in an organism or specific cell.
• Evolutionary Developmental Biology (EvoDevo) – Addressing the origins and evolution of embryonic development, this field (an outgrowth of the existing discipline of “Embryology”) has been empowered by data contained in genome sequencing of human And model organism cells.
• Computational Biology uses mathematical and computational approaches to address theoretical and experimental questions in biology. Its subdiscipline “computational genomics” perform statistical analysis on the data generated by gene sequencers and microarray technologies to evaluate gene and gene products expressed by various cell types
•Bioinformatics – Is perhaps best viewed as the applied twin of computational biology, and is well defined in the Sanger Institute’s glossary as the “science of using computer echnology to gather, store, analyze and merge biological data.Expertise in bioinformatics is key to handling the enormous amounts of data produced by the HGP and other sequencing projects, and serving it out to the researchers who use the data.”
• Metagenomics – Also known as environmental genomics or community genomics, meta genomics investigates the communal genome contained within an environmental sample. It enables the study of the symbiosis and interactions of organismal genomes, and genetic products as a biological system.
• Bioarchaeology and Anthropology – Uses genomics for the study of human evolution and population migrations. It has been noted that “comparative DNA sequence analyses of samples representing distinct modern populations of humans have revolutionized the field of anthropology.”
• Agricultural Sciences – Whole genome sequencing is being applied across a broad range of crop and livestock species and underpins genetic improvements in input and output traits. Genomics is also being used in diagnostics applications for livestock and zoonotic diseases, plant pathogen identification and in food safety applications. Work is also being performed in developing edible vaccines for incorporation into food products.
• Environmental Science – The U.S. Department of Energy and other investigators have sequenced the genomes of multiple nonpathogenic microbes useful for use and development In the environmental waste remediation, energy productions, carbon cycling and biotechnology applications.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT editor-in-chief@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
The fundamental advancement of scientific knowledge embodied in the human genome sequencing is also well illustrated by the broad variety of major biological science projects that have developed as follow‐on projects or outgrowths of the HGP. Table 11 lists some of these key related initiatives
Table 11 Building upon the human genome sequence related grand projects in biological sciences
|
PROJECT NAMES |
GOALS |
|
International Hap Map Project |
The Hap Map [17] is a public resource catalog of common genetic variants that occur in human beings. It describes what these variants are, where they occur in human DNA, and how they are distributed among people within populations and among populations in different parts of the world. Using the information in the Hap Map, researchers will be able to find genes that affect health, disease, an individual response to medications and environmental factors. |
|
1000 Genomes Project |
Identification of common genetic variation across humans through the Sequencing of a large number of individual human genomes. The project aims to find most genetic variants that have frequencies of at least 1 percent in the populations studied. |
|
ENCODE Project |
The National Human Genome Research Institute (NHGRI) launched a public research consortium named ENCODE, the Encyclopedia Of DNA Elements, in September 2003, to carry out a project to identify all functional elements in the human genome sequence. The project started with two components: a pilot phase and a technology development phase. The pilot phase tested and compared existing methods to rigorously analyze a defined portion of the human genome sequence. The conclusions from this pilot project were published in June 2007. The findings highlighted the success of the project to identify and characterize functional elements in the human genome. The technology development phase also has been a success with the promotion of several new technologies to generate high throughput data on functional elements. |
|
Cancer Genome Anatomy Project |
The Cancer Genome Anatomy Project (CGAP), begun in 1996, is an Interdisciplinary program established and administered by the U.S. National Cancer Institute to generate the information and technological tools needed to decipher the molecular anatomy of the cancer cell.
. |
|
Cancer Genome Atlas |
The Cancer Genome Atlas (TCGA) is a comprehensive and coordinated effort to accelerate our understanding of the molecular basis of cancer through the application of genome analysis technologies, including large‐scale genome sequencing. TCGA is a joint effort of the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI), which are both part of the National Institutes of Health, U.S. Department of Health and Human Services. |
|
Cancer Genome Project |
Headquartered at the Wellcome Trust Sanger Institute, the Cancer Genome Project is using the human genome sequence and high throughput mutation detection techniques to identify somatically acquired sequence variants/mutations and hence identify genes critical in the development of human cancers |
|
Human Microbiome Project |
Operated by the U.S. NIH Common Fund, the Human Microbiome Project (HMP) aims to characterize the microbial communities found at several different sites on the human body, including nasal passages, oral cavities, skin, gastrointestinal tract, and uro-genital tract, and to analyze the role of these microbes in human health and disease. |
|
Genomes to Life Project |
Genomes to Life program aims to use microbes and other organisms to address problems in energy production, environmental cleanup, and carbon cycling. The research seeks to understand the chemistry of entire organisms and their interactions with the environment. The project’s ten‐year goal is to advance systems biology, computation, and technology. These advances will be directed toward increasing biological‐based sources of energy, better understanding the earth’s carbon cycling, designing novel ways to capture carbon, and developing low‐cost methods for cleaning the environments. |
|
Microbial Genome Project[18] |
The MGP was begun in 1994 as a spinoff from the Human Genome Program. The program sequenced the genomes of a number of nonpathogenic microbes useful in solving DOE's mission challenges in environmental‐waste cleanup, energy production, carbon cycling, and biotechnology. |
BIOMEDICAL APPLIICATIONS OF HUMAN GENOME SCIENCE
HUMAN HEALTH AND MEDICINE - One of the key hopes for the sequencing of the human genome was that, having the entire genome in hand, biomedical scientists would be able to better identify the processes and mechanisms ofdisease and inherited medical disorders and illuminate new approaches to treating these diseases and disorders[19]. Modern medicine is built upon a history of fundamental investigations and discoveries across a broad range of life science disciplines: physiology, biochemistry, microbiology, immunology, etc. It is also, however, built upon a foundation of technological innovations enabling these discoveries to be made. Whether it is the invention of the microscope leading to the identification of microorganisms, or the modern development of functional magnetic resonance imaging allowing researchers to watch internal life processes in action, technologies provide new capabilities that propel medical advancement. The sequencing of the human genome actually represents an intense advance on both fronts both in terms of fun dam Human Health and Medicine. One ofthe key hopes for the sequencing of the human genome was that, having the entire genome in hand, biomedical scientists would be able to better identify the processes and mechanisms ofdisease and inherited medical disorders and illuminate new approaches to treating these diseases and disorders. Modern medicine is built upon a history of fundamental investigations and discoveries across a broad range of life science disciplines: physiology, biochemistry, microbiology, immunology, etc. It is also, however, built upon a foundation of technological innovations enabling these discoveries to be made. Whether it is the invention of the microscope leading to the identification of microorganisms, or the modern development of functional magnetic resonance imaging allowing researchers to watch internal life processes in action, technologies provide new capabilities that propel medical advancement. The sequencing of the human genome actually represents an intense advance on both frontental knowledge and technology advancement. As noted above, modern genomics and the human genome sequences have empowered a new view of biological processes. In turn, these new views are being translated into applied biomedical fields, particularly those illustrated in Figure 3
Fig-3: Functional impact areas of genomics in human health.
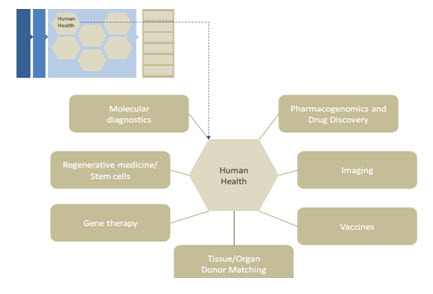
It is expected that the sequencing of human genome and the genomes of other organism will dramatically change our understanding and perception of biology and medicines. Some of the benefits of human genome project are given bellow in Table no-11
Table 11:
Biomedical applications of genomics and human genome sequencing [20]
|
Potential Application |
Genomics Advances Today |
Hope for the Future |
|
Diagnosis of single gene, Mendelian diseases and Disorders |
Specific genes for over 3,000 Mendelian monogenic diseases discovered. Genomic tests are being used to accurately diagnose rare diseases and disorders, many of which were previously misdiagnosed with inappropriate courses of treatment prescribed. Prenatal genetic screening is being performed to inform potential parents of risks for catastrophic inheritable disorders. |
Gene therapies will achieve success in repairing genetic abnormalities leading to diseases and disorders. Custom therapeutic products will block or change expressed activity of defective genes. |
|
Knowledge of predisposition towards specific diseases
|
Multiple genes and biomarkers have been identified for predisposition to Multiple diseases such as cancers. |
Understanding of risk for disease based upon multi‐gene tests will likely lead to appropriate therapeutic interventions and personal behavior/lifestyle modification
|
|
Genomics driven drug discovery, known as rational drug development |
New drug targets have been identified. Cancer drugs based on the genomics of tumors are on the market, including Gleevec (for chronic myelogenous leukemia), Herceptin (breastcancer), Tarceva (lung cancer) and Avastin (colon, lung and other cancers) |
Many new drugs and biologics will be developed to successfully exploit Elucidated drug targets. |
|
Therapeutic products custom prescribed based on patient genomics, to maximize effect and reduce or eliminate side effects |
Already being applied in the treatment of some forms of cancer and cardiovascular disease. Genetic tests are used for dosage levels in prescription of some drugs such as Coumadin (warfarin). |
Routine sequencing of a patient’s entire genome wil guide treatment selection and dose for the optimum response. Potential adverse reactions to drugs and treatment regimens, identified via genomic markers, will result in the avoidance of adverse events |
|
Repurposing or revitalization of some drugs shelved in development because of impact on a “genomic few” |
Successful discovery of subpopulations for which previously unapproved drugs are efficacious. Iressa, for example, has been approved with patents testing positive for the EGFR mutation |
There will be a substantial volume of existing drugs found to be efficacious in selected sub‐populations, and drug companies will have mined their previously “failed” R&D pipelines to bring forward previously nonmarketable drugs to work in selected sub‐populations. |
|
Identification of means to combat infectious organisms |
Multiple infectious organisms have had their whole genomes sequenced. Public health professionals sequence Emerging infectious disease organisms to monitor migrations and mutations. Genetic testing already being applied to direct therapy for HIV/AIDS patients. |
Rapid, real‐time sequencing of pathogens twill direct public health efforts to combat disease outbreaks in humans and zoonotic disease outbreaks in animals. DNA vaccines will come to the market to impart immunity to infectious diseases. Genetic profiling of patient viruses will assist in customizing Treatment regimens. |
|
Inherited genetic diseases and disorders |
After publicized setbacks, gene therapies are now achieving success. For example, the fatal brain disorder adrenoleuko dystrophy has been treated, with progression stopped, in a sample of children |
Gene therapy may be routinely provided to newborns with identified genomic profiles to correct defective genes, particularly in conditions associated with devastating monogenic disorders. |
Application of genomics to plant and livestock improvement—positively altering both organismal input traits (such as nutrient uptake, pest resistance, drought tolerance, etc.) and output traits (such as nutrient content, chemical composition, quality and quantity of food produced)[21]. Genomics is also being applied in the tracing of food contamination and associated pathogenic events. The benefits being felt in human healthcare also translate into veterinary medicine. The concurrent sequencing of various animal genomes during the HGP, and subsequent whole animal genome sequencing undertaken after completion of the human genome, has been as revolutionary for veterinary medicine as it has been for human biomedical sciences. Early in the HGP, the U.S. Department of Energy began applying genomics tools and technologies resulting from the human genome sequencing program to the sequencing of microbes for functionality in environmental remediation and for investigation of their use in other applications such as bioprocessing and biofuels production. The Microbial Genome Project and the recent Genomes to Life Project have specifically applied genomics to a range of environmental and industrial applications. Likewise commercial enterprises across a series of product categories in biotechnology, biofuels, food processing, drug and vitamin production, and biobased materials are applying advanced genomic knowledge and technologies to bring to market new and more efficient industrial processes to power the U.S. and global economies. Genomics has also become a tool for applications in the field of justice and security. For homeland security, the ability to genotype suspicious infectious pathogens and trace their origin is a national security priority. Law enforcement is also using genomics in identification of human remains in instances of natural disasters (such as the Haitian and Japanese earthquakes) or terrorist events (such as 9/11’s ground zero), and for tracing illegal trade in endangered animals. It would be impossible to quantify each and every impact being generated across the field of genomics and its application to such a broad range of scientific fields and technological disciplines. Battelle has not attempted to do that. Rather Battelle examined each of the key fields of application shown in Figure -1 and discusses within the full report the ways in which genomics is being applied within these fields, and provides case studies of genomics in action within these fields.
FUTURE OF HUMAN GENOME SCIENCE :-- The advancement of knowledge and the technologies resulting from human genome sequencing have formed the platform for nothing less than a medical revolution[22]. The primary impacts of this revolution in quantitative and personalized medicine may not yet be felt in daily clinical practice (although in some areas of cancer it is), but that day is accelerating towards us. One of the key realizations that must be understood regarding the human genome sequencing is that its usefulness is perpetual. While other major big science projects have a life attached to them the human genome sequence will not wear out or become obsolete.4 Rather, the reference human genome is akin to chemistry’s periodic table, a perpetually useful fundamental platform for understanding and advancing science. The impact on human medicine and health is profound and important but, as shown, the benefits of performing the HGP and related projects extend into areas of far beyond human health. Our fundamental understanding of genomics, the sequencing and genomic technologies whose development was spurred by the HGP, and the advanced “omics” disciplines created through these efforts, will continue to make contributions on a broad range of fronts. We can expect a variety of genomics‐enabled enhancements, for example: 1. Agricultural productivity to increase considerably, working towards the challenge of feeding the world’s rapidly expanding population in a sustainable manner.
2. Not only will food availability increase, but the impact of its production on the global environment will reduce as crops and livestock are developed with traits suited to nitrogen Use efficiency, no‐till agriculture, water use efficiency and reduced waste production.
3. Currently low‐value biomass, especially low‐value cellulosic biomass, will be converted Into higher‐value liquid fuels, energy sources, biobased chemicals, plastics and materials. These products will increasingly displace petroleum and other fossil‐based inputs, contributing to reduced carbon emissions and associated climate and environmental benefits.
4. An increasingly two‐way flow of diagnostics, therapeutics and prevention tools will move Between human medicine, veterinary medicine and agriculture as the cost of genomic technologies reduces and the applications of discoveries in one area can be applied to another because of comparative genomics and other genomic advancements.
5. The legacy of pollution on the planet caused by human activity will be addressed increasingly through the application of genetically engineered, modified or synthetic organisms designed to perform remediation and mitigation functions
6. The biochemistry and genetics of much single cell disorder have been elucidated e.g.sickle cell anemia, cystic fibrosis, and retinoblastoma. A majority of common disease in humans are polygenic e.g. cancer hypertension and diabetes .At present we have a very little knowledge about the cause of the disease. The information on the genome sequence will certainly help to unravel the mysteries surrounding the polygenic disease
7. The drug may be tailored to treat individual patient. This will become possible considering the variation in enzyme and other protein involved in drug action.
8. With the genome sequence now in hand the complex social traits can be better understood .For instance gene controlling speech have been identified 9.Genomes of many organisms have been sequenced and this number will increased in recent years .The information on genome of different species will through light on major stages on evolution.
10. At present human genome therapy is in its infancy due to various reasons .Genome sequence knowledge will certainly help for more effective treatment of genetic disease.
The sequencing of the human genome was the signature scientific program of the 1990’s and early 2000’s. When Science, in 2010, produced a retrospective on the ten most important insights (Table-12) of the decade, five of the ten most influential scientific insights stem from, or make use of, the human genome sequencing and related genomics advances.
Table 12: Science magazine, top insights of the decade 2010[23]
|
INSIGHT |
DESCRIPTION |
|
Genomic Complexity |
“Gene regulation has turned out to be a surprisingly complex process governed by various types of regulatory DNA, which may lie deep in the wilderness of so‐called junk DNA that lies between genes. Far from being humble messengers, RNAs of all shapes and sizes are actually powerful players in how genomes operate. Finally, there's been increasing recognition of the widespread role of chemical alterations called epigenetic factors that can influence the genome across generations without changing the DNA sequence itself. The scope of this “dark genome” became apparent in 2001, when the human genome sequence was first published. |
|
The Human Micro biome |
This past decade has seen a shift in how we see the microbes and viruses in and on our bodies. There is increasing acceptance that they are us, and for good reason. Nine in 10 of the cells in the body are microbial. In the gut alone, as many as 1,000 species bring to the body 100 times as many genes as our own DNA carries. A few microbes make us sick, but most are commensal and just call the human body home. Collectively, they are known as the human microbiome. Likewise, some viruses take up residence in the body, creating a virome whose influence on health and disease is just beginning to be studied. |
|
Stem Cells and Regenerative Medicine |
By prompting a cell to over express a few genes, researchers have discovered in the past decade how to turn a skin or blood cell into a pluripotent cell: one that has regained the potential to become any number of cells in the body. Other genes can prompt skin cells to turn directly into neurons or blood cells. Scientists are already using the technique to make cell lines from patients with hard to study diseases, and ultimately they hope to grow genetically matched replacement cells and tissues—perhaps even entire organs. |
|
Ancient DNA |
In the past decade, powerful new x‐ray scans and three‐dimensional computer models have transformed the analysis of ancient bones, teeth, and shells. But a new kind of analysis is capable of revealing anatomical adaptations that skeletal evidence can't provide, such as the color of a dinosaur's feathers or how woolly mammoths withstood the cold. The new views of the prehistoric world hinge on the realization that “biomolecules” such as ancient DNA and collagen can survive For tens of thousands of years and give important information about long‐dead plants, animals, and humans. |
THE FUTURE OF GENOMICS IN MEDICINE[24] :
DIAGNOSTICS- Appropriate treatment of a disease can only begin once an accurate diagnosis is made. Genomics will increasingly drive advanced, highly accurate diagnostics development by:
1. Identification of variant genes and their impact on monogenic and mutagenic diseases.
2. Sub classification of diseases by genome analysis to direct personalized medicine approaches (as is already occurring in certain cancers).
3. Methods for real‐time pathogen detection and identification.
THERAPEUTICS-Genomics will significantly increase in utility across a range of therapeutic R&D and application areas, including:
1. Identification of specific disease‐relevant targets.
2. Rational drug design to produce drugs specifically aimed at the characteristics of the identified targets.
3. Repurposing of existing drugs for application to newly identified genomic targets and signatures.
4. Previously failed drugs “rescued” for application to genomic sub‐populations.
5. Clinical trials improved through genomic stratification of participants significantly reduced adverse drug events, and associated costs, driven by genotype‐guided drug prescription.
6. Increasing application of gene therapies.
7. Therapeutic strategies developed for genotype environment interactions and for adaptation to
behavioral and lifestyle characteristics.
ETHICS AND HUMAN GENOME:-
The research on human genome science will make every sensitive data available that will affect the personal and private lives of individual. For instance, once it is known that a person carries a gene for an incurable disease, what would be the strategy of an insurance company and how will the society treat him /her? That is a possibility that individual with substandard genome will be discriminated .Human genome results may also promote racial discrimination categorizing the people with good and bad genome sequence .Considering the gravity of ethical related to human genome about 3% of total HGP budget was earmarked for ethical research[25].
CONCLUSION:
As the Human Genome Project (HGP) continues toward its goal to identify each of the 3 billion base pairs that make up the human genome, some believe the solution to the mysteries of humanity, the most magnificent and complex biological Rosetta stone may be near. The seemingly disparate paths of the sciences of molecular biology, chemistry, and physics have converged to this end. The work of the HGP and the knowledge it spawns will someday reveal
a new human anatomy. As James Watson has recognized, “if society is to cope with the consequences of this knowledge, people must learn and become better informed about genetics.” Proponents of the HGP view it as a trustworthy and badly needed step toward improving the health of mankind, but ethical debates must acknowledge the horrors perpetrated in the name of eugenics in the past century. The important dialogue already begun must continue with a focus on forestalling the threat of creating new forms of discrimination and methods of oppression. Much of the suspicion of the HGP is a presumed hidden agenda to control the future of humanity through manipulating human genes and the reductionist philosophy that humans are “determined” by their genes. Almost everyone would agree that, for the foreseeable future, new genetic knowledge should be directed toward its use to prevent or cure disease. On the other hand, is there anything wrong with pursuing perfection as a goal of reproduction? Will it be part of medicine’s professional responsibility to allow parents to select among traits for their future offspring? Will such choices only be available to those who are able to pay? Without serious considerations of these difficult questions today, society of the future risks being divided into those who are genetically sound and those who are genetically afflicted. There are serious inequalities in the United States in access to medical services which are correlated to both class and race. A world in which genetic testing is widely available might be incompatible with a system of private health insurance. Societies committed to equality and fairness will have to consider measures to ensure that the means to implementing eugenic choices are available to all who desire them. Envisaging the future world of genomics reinforces arguments for ensuring all members of society will have access to affordable medical care, so that the new technologies do not compromise the rights and aspirations of vulnerable people. The USD 3.8 billion spent on the HGP may well represent the best single investment ever made in science.
Human Genome Project is one of those achievements of the human spirit that
Make me proud to be human.- Richard Dawkins It is humbling for me and awe inspiring to realize that we have caught the first glimpse Of our own instruction book, previously known only to God- Francis Collins
REFERENCE:
1. DeLisi, Charles (1988). "The Human Genome Project". American Scientist 76: 488
2. U.Satyanarayana.et.al “Biotechnology” 2nd Edn 2012, 148-149.
3. DeLisi, Charles (1988). "The Human Genome Project". American Scientist 76: 488. Bibcode:1988AmSci..76.488D
4. International Human Genome Sequencing Consortium (2001). "Initial sequencing and analysis of the human genome" (PDF). Nature 409 (6822): 860–921.
5. Noble, Ivan (2003-04-14). "Human genome finally complete". BBC News. Retrieved 2006-07-22.
6. Davidson College (2002). "Sequencing Whole Genomes: Hierarchical Shotgun Sequencing v. Shotgun Sequencing". Bio.davidson.edu. Department of Biology, Davidson College. Retrieved 1 August 2013.
7. Osoegawa, Kazutoyo; Mammoser, AG; Wu, C; Frengen, E; Zeng, C; Catanese, JJ; De Jong, PJ (2001). "A Bacterial Artificial Chromosome Library for Sequencing the Complete Human Genome". Genome Research 11 (3): 483–96.
8. Clamp, M. et al. 2007. “Distinguishing protein‐coding and noncoding genes in the human genome.” Proceedings ofthe National Academy of Sciences USA. 104. 2007.
9. Stewart Scherer. 2008. “A Short Guide to the Human Genome.” Section on how many protein‐coding genes arepresent in the genome. Cold Spring Harbor Laboratory Press, New York.
10. Harmon, Katherine (2010-06-28). "Genome Sequencing for the Rest of Us". Scientific American. Retrieved 2010-08-13.
11. Venter D (2003). "A Part of the Human Genome Sequence". Science 299 (5610): 1183–4.
12. Eric S. Lander. 2011. “Initial impact of the sequencing of the human genome.” Nature. Volume 470, February 10, 2011.
13. J. D. Watson, “The human genome project: Past, Present, and Future,” Science, vol248, pp. 44–48, 1990.
14. Bryant, J. A (2007). Design and information in biology: From molecules to systems. p. 108. ISBN 9781853128530. "...brought to light about 1200 protein families. Only 94 protein families, or 7%, appear to be vertebrate specific.
15. Lander ES (2011). "Initial impact of the sequencing of the human genome". Nature 479 (7333): 187–197.
16. C. R. Cantor, “Orchestrating the human genome project,” Science, vol. 248, pp. 49–51, 1990.
17. Naidoo N, Pawitan Y, Soong R, Cooper DN, Ku CS (2011). "Human genetics and genomics a decade after the release of the draft sequence of the human genome". Hum Genomics 5 (6):577 –622.
18. Sun‐Hee Hong, et al. 2006. “Predicting Microbial Species Richness.” Proceedings of the National Academy of Sciences of the United States (PNAS). January 3, 2006. Volume 103, Number 1. Pages 117‐122.
19. M. L. Pearson, and D. Soll, “The human genome project: A paradigm for information management in the life sciences, The FASEB Journal, vol 5, pp. 35–39, 1991.
20. George E. Liu. 2009. “Applications and case Studies of the Next‐Generation Sequencing Technologies in Food, Nutrition and Agriculture.” Recent Patents on Food, Nutrition and Agriculture, 2009. 1, 75‐79.
21. Snyder M, Du J, Gerstein M (2012). "Personal genome sequencing: current approaches and challenges". Genes Dev 24 (5): 423–431.
22. Gonzaga-Jauregui C, Lupski JR, Gibbs RA (2012). "Human genome sequencing in health and disease". Annu Rev Med 63 (1): 35–61
23. Human Genome Project Information Archive (2013). "U.S. & International HGP Research Sites". U.S. Department of Energy & Human Genome Project. Retrieved 1 August 2013.
24.Leroy Hood. 2002. “After the Genome: Where Should We Go?” Chapter in Michael Yudell and Rob DeSalle (editors) “The Genomic Revolution: Unveiling the Unity of Life.” 2002. Joseph Henry Press, Washington DC with the American Museum of Natural.
25. Greely, Henry (1992). The Code of Codes: Scientific and Social Issues in the Human Genome Project. Cambridge, Massachusetts: Harvard University Press. pp. 264–65.
|
PharmaTutor (Print-ISSN: 2394 - 6679; e-ISSN: 2347 - 7881) Volume 5, Issue 10 Received On: 07/06/2017; Accepted On: 27/06/2017; Published On: 01/10/2017 How to cite this article: Sameena T, Patil P, Sethy SP; Human Genome Science: A new face of pharmaceutical science: A Review; PharmaTutor; 2017; 5(10); 30-47 |
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT editor-in-chief@pharmatutor.org
FIND OUT MORE ARTICLES AT OUR DATABASE









