About Author:
Mariz Sintaha
Lecturer,
School of Life Sciences
Independent University, Bangladesh
sintu.bmb@gmail.com
Introduction:
MRSA refers to strain of Staphylococcus aureus which is resistant to methicillin. MRSA infection causes Red bump (may be pus filled), warmth, pain, swollen and red and tender skin lesions and are resistant to various antibiotics used for treatment including cephalosporins and penicillins. Rapid and accurate detection of MRSA has great significance since is needed for proper treatment of staph diseases. It is difficult to detect MRSA in media using phenotypic methods such as disk diffusion, oxacillin agar screening test, agar dilution, MIC determination by broth dilutionsince expression of metcillin resistance is not uniform in all cells. The genotypic method is now taken as the gold standard to detect MSRA because of its accuracy.
Reference Id: PHARMATUTOR-ART-1969
Polymerase Chain Reaction (PCR)
Principle- Polymerase chain reaction is a technique widely used in field of Molecular Biology for various purposes. The technique is used to amplify a specific segment of DNA across several orders of magnitude. The DNA to be amplified is mixed with all the materials needed for synthesizing new DNA strand and a pair of primer to specify the region of DNA to be amplified. Temperature of the mixture is fixed into three steps one after another- the denaturation temperature (94oC), the annealing temperature (55oC to 65oC) and primer extension temperature (72oC) (William B. Coleman 2006). These three steps constitute a cycle. In each cycle the copy number of duplex DNA doubles (theoretically). The cycles are repeated for many times by the aid of an automated temperature regulating machine “Thermocycler”. After few cycles, size of the newly synthesized DNA is fixed as the distance between 5’ ends of primers (Lo 1998). Thus, at the end of the reaction (after completion of many cycles) millions copies of specific segment of template DNA are obtained.
Reagents- The PCR mixture contains (William B. Coleman 2006)-
1. Heat resistant DNA synthesizing enzyme- Taq polymerase, collected from the bacteria T. aquaticus.
2.Two short segments of DNA (18-30 bp long) named forward and reverse primer, that is complementary to the 3’ ends of the region of DNA to be amplified of the sense and anti-sense strand respectively.
3. Deoxy nucleotides triphosphates (dTTP, dGTP, dCTP and dTTP), which are building blocks of DNA
4. Tris-Cl buffer of PH 8.3 in which taq polymerase shows maximum activity.
5. Divalent ions, such as Mg+ which is required for dissociation Taq polymerase from DNA.
6. KCl which promotes primer annealing to template DNA.
7. Template DNA
Mechanism- In denaturation step, high temperature causes melting of DNA from double strand to single strand. Generally in first cycle a higher denaturation temperature is used to cause complete melting of duplex DNA and activation of taq polymerase which remains in inactivated state in commercially available solution. A Lower annealing temperature allows primers to bind with the separated single stranded DNAs. At primer extension temperature, Taq polymerase starts adding dNTPs at the 5’ end of the primer complimentary to the template strand. Thus a new DNA strand is synthesized which is complimentary to the template. The product of first cycle provides template for second cycle. After completion of all cycles, often temperature is hold at 70-74o C for 7-10 minutes so that remaining single stranded DNA can be extended and then hold at 4-15o C until it is removed for further work (Michael A. Innis 1999).
Visualization of PCR Product- For visualization, PCR product is resolved through Gel Electrophoresis. Million copies of similar sized DNA produced by PCR, concentrate in a location of the gel. The gel is then stained with a fluorescent dye “ethidium bromide”. The dye intercalates in to the duplex DNA. After washing the gel with distilled water to remove unbound ethidium bromide, DNA band is visualized under UV light (Mathews n.d.).
Methicillin-resistant Staphylococcus aureus (MRSA)
Staphylococcus aureus (S. aureus) is a facultative pathogenic Gram-positive bacterium that inhabits in human skin and mucosa. It causes disease occasionally by getting into the body. The causes disease in virtually any tissue diseases including wound, skin and urinary tract infection, pneumonia, sepsis and osteomyelitis (Chambers 2009). These diseases can be treated successfully by beta-lactum antibiotics since most strains of Staphylococcus aureus are sensitive to it. But the strains resistant to methicillin (Methicillin-resistant Staphylococcus aureus or MRSA) are resistant to various antibiotics used for treatment including cephalosporins and penicillins. Clinical use of methicillin is responsible of increasing the incidence of acquiring methicillin resistance by Staphylococcus aureus(MRSA). Other factors responsible include use of intravenous drug, uncontrolled use of antibiotics, nose infection of Methicillin-resistant Staphylococcus aureus (MRSA)and prolonged stay in hospital (Lu PL 2005). Methicillin-resistant Staphylococcus aureus (MRSA)is the major cause of community acquired and hospital-acquired infection in recent decades (Martins 2007). The symptoms of MRSAinfection include Red bump (may be pus filled), warmth, pain, swollen and red and tender skin lesions.
Polymerase Chain Reaction (PCR) to identify MRSA
Methicillin-resistant Staphylococcus aureus (MRSA) causes serious infection that is difficult to treat. For effective treatment and prevent spread of the disease, Methicillin-resistant Staphylococcus aureus must be identified in clinical specimen rapidly and accurately. Methcillin resistance occurs in Staphylococcus aureusby acquiring a gene called MecA (Hartman BJ 1984). This gene encodes an altered form of penicillin binding protein which has low affinity for beta lactum antibiotics (Lim 2002). Thus the presence of a gene in the DNA isolated from clinical specimen is the indicator of the Methicillin-resistant Staphylococcus aureus infection. Polymerase chain reaction is used widely to identify a specific DNA sequence within a sample. Since, polymerase chain reaction involves binding of primers complimentary to specific sequences of DNA and amplifying a specific segment of DNA, successful polymerase chain reaction proves the presence of a specific DNA sequence. MecA gene is conserved in Staphylococcus species, so detecting MecA by PCR is considered as standard method for identifying Methicillin-resistant Staphylococcus aureus (MRSA) (Maes N 2002), (Bignardi GE 1996) (Murakami K 1991).
Actually, the resistant bacteria acquires a transposable element named staphylococcal cassette chromosome (SCCmec) in which the MecA gene is contained (C. Cuny 2005). Sccmec contains incomplete inverted repeat at both ends and integrated at the 3′-end of orfXregion, within a site named attBSCC (Hiramatsu K 2002). SCCmec element is classified in five classes according to the mec gene, insertion complex, site-specific recombinase genes and transposon tn554 (Ma X 2002). Identification of MecA gene alone can give false positive result since it is present both in Methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci. So polymerse chain identification of the region covering theorfX and SCCmec can be used to specifically identify Methicillin-resistant Staphylococcus aureus. For these purpose, five forward primers for five types of SCCmec elements and one reverse primer fororfX region can be used (Huletsky A 2004).
Coagulase-negative staphylococci (CoNS) is another vital cause of infection (von Eiff C 2001) and commonly isolated in laboratories (CC. 1990). Coagulase-negative staphylococci (CoNS) also contain multi-drug resistance. So MRSA and CoNS must be distinguished (Zhang K 2004) for rapid detection and proper treatment ofMRSA. MRSA and CoNs are not distinguishable in PCR detection method of MecA gene since MecA genes of both species have 99% sequence similarity (Ryffel C 1990). For this purpose presence coag gene (S Rallapalli1 2008)is analysed besides MecA gene in multiplex PCR systems. In MRSA both MecA and coag genes are detected by PCR where in CoNS, only MecA gene is detected by PCR (S Rallapalli 2008). Another method of distinguishing MRSA and CoNS is multiplex PCR to co-amplify both mecA gene responsible for methcillin resistance and femA gene responsible for coagulase activity (Kohner P 1999) (Sakoulas G 2001) (Vannuffel P 1995) (Kobayashi N 1994) since mecA gene is present both in MSRA and CoNS where femA is present only is MSRA.
PCR methodologies
|
|
Forward Primer 5’-3’ direction |
Reverse Primer 5’-3’ direction |
Reaction Mix |
Cycles |
Length (bp) |
Reference |
|
SCCmec elements |
rjmec 57641–57656 TATGATATGCTTCTCC |
ORFX1r 58042–58022 AACGTTTAGG |
Total Volume= 25 µL
-Taq DNA polymerase (1.3 U) -0.5 µL DNA (10 ng) -primer(2.5 pmol) -dNTP(200 µM) each in Tris-HCl (10 mM) -KCl(500 mM) -MgCl2(1.5 mM)
|
Total 30 cycles
Initial Denaturation 94°C 3 min
Denaruration- 94°C 30 s
Annealing- 55°C 30 s
Extension- 72°C 30 s
Final extension- 72°C 4 min. |
400 |
(C. Cuny 2005) |
|
mecA |
mecA1 318-338 GTAGAAATGACT |
mecA2 603-627 CCAATTCCACATT |
Total volume=50ml
-Taq DNApolymerase(2U) -DNA(30 ng) -each primer(50 pmol) -dNTP(200 mM) -10X reaction buffer[(NH4)2SO4((160 mM), Tris HCl (670 mM ) of pH8.8, Tween 80( o.1%)] -MgCl2(2.5 mM)
|
Total 30 cycles Initial denaturation 94o C five minutes, denaturation a 94oC 30 seconds annealing 55oC 30 seconds Extension 72oC 30 seconds final extension 72oC 10 minutes. |
310 |
(Riza Adaleti 2008) |
|
mecA |
primer 1 37-57 GTTGTAGTT |
Primer 2 1827-1854 CCACCCAATlTTGTCT |
Total Volume=25 μL
-Taq polymerase (2.5 U) -3 μL of template DNA -Primers (100 pmol of each) -dNTPs -PCR Buffer Mg (2.5 mm) |
Total 40 cycles
Initial denaturation 94o C 3 minutes
Denaturation 94 °C 30 sec
Annealing 60 °C 45 sec
Extension 72 °C 1.5 minutes
Final Extension 72 °C 3.5 minutes. |
1800 |
(S Ercis 2010) |
|
Multiplex PCR to Distinguish Between MRSA and CoNS |
||||||
|
MecA
coag |
1282 to 1303 AAAATCGATG
CGAGACCAA |
1793 to 1814 AGTTCT
AAA GAA AAC |
Total Volume=20 µL
- 1µL DNA (50 ng) -Buffer (500mM KCl, 100mM Tris-HCl, pH 8·3) - dNTP(0.5 mM each) - MgCl 2 (2.5 mM) -coag primer (0.15 µM of each ) -mecA primer(0.1 µM) - Taq DNA polymerase(2U)
|
Total 40 cycles
Initial denaturation 95o C five minutes,
Denaturation a 94oC 30 seconds
Annealing 55oC 30 seconds
Extension 72oC 90 seconds
Final extension 72oC 5 minutes.
|
533
810 |
(S Rallapalli 2008) |
|
mecA
femA |
MecAl 1282-1303 AAAATCGATGG
FemAl 595-614 AGACAAATA |
MecA2 1739-1814 AGTTCTGCA
FemA2 1084-1103 AAATCTAA |
Total Volume= 100 µL - Taq DNApolymerase (1U) -10 µL DNA -Primer (100 pM each) -dNTP (200 µM each) -Tris-HCl (10 mM, pH 8 3), -KCI(50 mM) -MgCl2(1-5 mM) |
Total 25 cycles
Denaturation 94oC 1 minutes
Annealing 57oC 1 minutes
Extension 72oC 2 minutes
Final extension 72oC 5 minutes.
|
533
509 |
(Kobayashi N 1994) |
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
Reliability and Accuracy of PCR compared to other techniques
Rapid and accurate detection of MRSA has great significance since is needed for proper treatment of staph diseases (disease caused by Staphylococcus) (HF 1993). Vitek automatic system, disk diffusion, oxacillin agar screening test,agar dilution, MIC determination by broth dilution are the phenotypic methods used widely to detect Methicillin-resistant Staphylococcus aureus (Frebourg NB 1998)(Sakoulas G 2001). But it is difficult to detect Methicillin-resistant Staphylococcus aureus by phenotypic method since the cells which acquired mecA gene does not express metcillin resistance uniformly (Hartman BJ 158:513-6) and actually, only a few cells express metcillin resistance (Matthews PR 1984). Some other factors make it more complex to detect Methicillin-resistant Staphylococcus aureus such as pH of the medium, incubation time and temperature, inoculum size, exposure to beta lactam antibiotics and salt concentration of the medium since they all determines the degree of expression of metcillin resistance (DF. 2001). Borderline oxacillin resistant Staphylococcusaureus (BORSA) and modified Staphylococcusaureus (MODSA) show oxacillin resistance by producing high concentration of beta lactamase and a modified penicillin binding protein rather than acquiring new penicillin binding protein respectively. None of them contain the mecA gene. So, BORSA and MODSA may cause false negative result by phenotypic methods during detection of Methicillin-resistant Staphylococcus aureus (Anila A Mathews 2010).The genotypic method is proved as more accurate than phenotypic method by study (Chambers 2009) (Frank Kipp 2004) as well as it is devoid of all the limitations of phenotypic method. That’s why it is taken as gold standard for identifying Methicillin-resistant Staphylococcus aureus (Sakoulas G 2001) (Unal S 1994).
Labeled diagrams
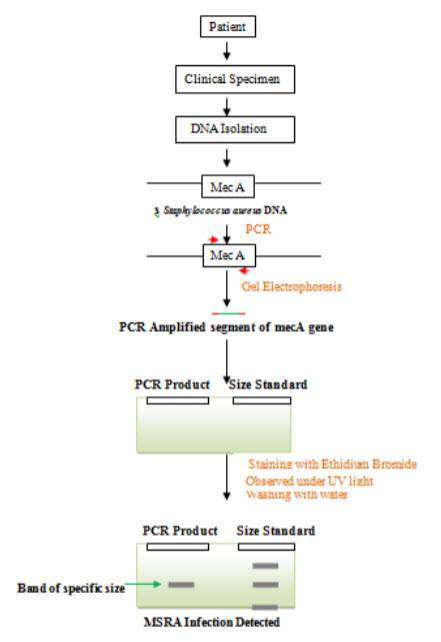
Figure- Detection of MSRA by PCR
Reference
- Anila A Mathews, MTBAJJ 2010, 'Evaluation and comparison of tests to detect methicillin resistant S. aureus', Indian j of Patho. and Microbiol., vol 53, no. 1, pp. 79-82.
- Bignardi GE, WNCAJASD 1996, ' Detection of the mecA gene and phenotypic detection of resistance in Staphylococcus aureus isolates with borderline or low-level methicillin resistance.', J Antimicrob Chemother, vol 37, pp. 53-63.
- C. Cuny, WW 2005, 'PCR for the identification of methicillin-resistant Staphylococcus aureus (MRSA) strains using a single primer pair specific for SCCmec elements and the neighbouring chromosome-borne orfX', Clinical Microbiology and Infection, vol 11, no. 10, pp. 834-837. CC., P 1990, ' Coagulase-negative staphylococci: pathogens with increasing clinical significance.', J Pediatr. , vol 116, no. 4, p. 497–507.
- Chambers, HF,AFRD 2009, 'Waves of resistance: Staphylococcus aureus in the antibiotic era.', Nat. Rev. Microbiol., vol 7, pp. 629-41.
- DF., B 2001, 'Detection of methicillin/oxacillin resistance in staphylococci.', J Antimicrob Chemothe, vol 48, pp. 65-70.
- Frank Kipp, KBGPACVE 2004, 'Evaluation of Different Methods To Detect Methicillin Resistance in Small-Colony Variants of Staphylococcus aureus', J Clin Microbiol., vol 42, no. 3, p. 1277–1279.
- Frebourg NB, NDLLMELJ 1998, 'Comparison of ATB Staph, Rapid ATB Staph, Vitek and Etest methods for detection of oxacillin heteroresistance in staphylococci possessing mecA.', J Clin Microbiol, vol 36, pp. 52-57.
- Hartman BJ, TA 158:513-6, 'Low-affinity penicillin-binding protein associated with β -lactam resistance in Staphylococcus aureus ', J Bacteriol, vol 158, p. 1984.
- Hartman BJ, TA 1984, 'Tomasz A. Low-affinity penicillin-binding protein associated with β -lactam resistance in Staphylococcus aureus. ', J Bacteriol , vol 158, pp. 513-6.
- HF, C 1993, 'Detection of methicillin-resistant staphylococci.', Infect Dis Clin North Am , vol 7, pp. 425-33.
- Hiramatsu K, KYYHIT 2002, 'Molecular genetics of methicillin-resistant Staphylococcus aureus. ', Int J Med Microbiol, vol 292, p. 67–74.
- Huletsky A, GRRVEA 2004, 'New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci.', J Clin Microbiol , vol 42, p. 1875–1884.
- Kobayashi N, WHKKTKUSUNEA 1994, 'Detection of mecA, femA and femB genes in clinical strains of staphylococci using polymerase chain reaction.', Epidemiol Infect, vol 113, pp. 259-66.
- Kohner P, UJKCPDCIF 1999, 'Comparison of susceptibility testing methods with mec-A gene analysis for determining methicillin resistance in clinical isolates of Staphylococcus aureus and coagulase negative Staphylococcus spp.', J Clin Microbiol, vol 37, pp. 2952-61.
- Lim, D,ANCS 2002, 'Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus.', Nat. Struct. Biol., vol 9, pp. 870-876.
- Lo, YMD 1998, Clinical Applications of PCR, Humana Press Inc. , Totowa, New Jersey.
- Lu PL, CLPCCYCTMLEA 2005, 'Risk factors and molecular analysis of community methicillin-resistant Staphylococcus aureus carriage', J Clin Microbiol, vol 43, pp. 132-9.
- Ma X, ITTCEA 2002, 'Novel type of staphylococcal cassette chromosome mec identified in community acquired methicillin-resistant Staphylococcus aureus. ', Antimicrob. Agents. Chemoth., vol 46, p. 1147–1152.
- Maes N, MJRSDGYSM 2002, 'Evaluation of a triplex PCR assay to discriminate Staphylococcus aureus from coagulase-negative staphylococci and determine methicillin resistance from blood cultures.', J Clin Microbiol , vol 40, pp. 1514-7.
- Martins, A,ALCM 2007, 'Methicillin resistance in Staphylococcus aureus and coagulase-negative staphylococci: epidemiological and molecular aspects.', Microbiol. Immunol., vol 51, pp. 787-795.
- Mathews, VHA, Biochemistry, 3rd edn, Pearson Education Inc. Matthews PR, SP 1984, 'Resistance heterogeneity in methicillin-resistant Staphylococcus aureus ', FEMS Microbiol Lett , vol 22, pp. 161-6.
- Michael A. Innis, JJSDHG 1999, PCR applications: protocols for functional genomics, Academic Press, SanDiego, California, USA.
- Murakami K, MWWKNETHWS 1991, 'Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. ', J Clin Microbiol , vol 29, pp. 2240-4.
- Riza Adaleti, 1YNZCKCTFK1 2008, 'Comparison of polymerase chain reaction and conventional methods in detecting methicillin-resistant Staphylococcus aureus', J Infect Developing Countries , vol 2, no. 1, pp. 46-50.
- Ryffel C, TWB-MIRPB-MLFB-BB 1990, 'Sequence comparison of mecA genes isolated from methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis.', Gene, vol 94, no. 1, pp. 137-138.
- S Ercis, BSGH 2010, 'A comparison of PCR detection of Meca with oxacillin disk susceptibility testing in different media and sceptor automated system for both Staphylococcus aureus and coagulase-negative staphylococci isolates', Indian Journal of pathology & Microbiology, vol 53, no. 1, pp. 79-82.
- S Rallapalli, SVRV 2008, 'Validation of multiplex PCR strategy for simultaneous detection and identification of methicillin resistant Staphylococcus aureus', Indian Journal of Medical Microbiologists , vol 26, no. 4, pp. 361-364.
- S Rallapalli1, SVRV 2008, 'Validation of multiplex PCR strategy for simultaneous detection and identification of methicillin resistant Staphylococcus aureus', Indian Journal of Medical Microbiologists, vol 26, no. 4, pp. 361-364.
- Sakoulas G, GHVLDPEGQQ 2001, 'Methicillin resistant Staphylococcus aureus: Comparison of susceptibility testing methods and analysis of mecA positive susceptible strains', J Clin Microbiol, vol 39, pp. 3946-51.
- Sakoulas G, GHVLDPEGQQ 2001, ' Methicillin-resistant Staphylococcus aureus: Comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. ', J Clin Microbiol, vol 39, pp. 3946-51.
- Unal S, WKDPBFEG 1994, 'Comparison of tests for detection of methicillin resistant Staphylococcus aureus in a clinical microbiology laboratory', Antimicrob Agents Chemother, vol 38, pp. 345-7.
- Vannuffel P, GJEHVDCBDMWGEA 1995, 'Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR.', J Clin Microbiol, vol 33, pp. 2864-7. von Eiff C, PRPGC-NS 2001, 'Pathogens have major role in nosocomial infections', Postgrad Med. , vol 110, no. 4, p. 63–64.
- William B. Coleman, GJT 2006, Molecular Diagnostics for The Clinical Laboratorian, 2nd edn, Humana Press, New York.
- Zhang K, SJCBESHZCDGDLTCJ 2004, 'New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci.', J Clin Microbiol., vol 42, no. 11, p. 4947–4955.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
FIND OUT MORE ARTICLES AT OUR DATABASE









