About Authors:
Avisek Mukhopadhyay*, Subhasis Banerjee
Gupta College Of Technological Sciences
M.Pharm(Bit Mesra, Ranchi)
*avisekmukhopadhyay7@gmail.com
INTRODUCTION
Urbanization, industrialization, changes in lifestyles, population growth and ageing all have contributed for epidemiological transition in the country. The absolute number of new cancer cases is increasing rapidly, due to growth in size of the population, and increase in the proportion of elderly persons as a result of improved life expectancy following control of communicable diseases. In India, the life expectancy at birth has steadily risen from 45 years in 1971 to 62 years in 1991, indicating a shift in demographic profile. It is estimated that life expectancy of Indian population will increase to 70 years by 2021–25. Such changes in the age structure would automatically alter the disease pattern associated with ageing and increase the burden of problems such as cancer, cardiovascular and other non-communicable diseases in the society.The latest global figures show that in 2004, there were 11.4 million cases of cancer diagnosed worldwide, and 75% of these were in Europe, the Americas and the Western Pacific Region. The Western Pacific Region includes China, Malaysia, Japan, Australia and New Zealand. The lowest numbers of cases were diagnosed in the Eastern Mediterranean region (includes Saudi Arabia, Egypt Iraq, Morocco, Tunisia) and Africa.
There were differences in the top three most common cancers in each region. The top three included lung cancer in all regions except Africa and included breast cancer in all except for the Western Pacific region. Cervical cancer was in the top three in Africa and South-East Asia only and bowel cancer in the top three for Europe only. Prostate cancer was in the top three in Africa and the Americas.Let us discuss one of the most promising targets of these days, which when overexpressed may cause cancer, i.e.;HISTONE DEACETYLASE.
[adsense:336x280:8701650588]
Reference Id: PHARMATUTOR-ART-1425
ROLE OF HISTONE DEACETYLASE IN HUMAN CANCER
Over the past few years, a lot of efforts have been done in the field of Histone deacetylase inhibitors (HDACi) and more than a hundred patents claiming new chemical series have emerged.The balance of histone acetylation and deacetylation is an epigenetic layer with a critical role in the regulation of gene expression. Histone acetylation induced by histone acetyl transferases (HATs) is associated with gene transcription, while histone hypoacetylation induced by histone deacetylase (HDAC) activity is associated with gene silencing. Altered expression and mutations of genes that encode HDACs have been linked to tumor development since they both induce the aberrant transcription of key genes regulating important cellular functions such as cell proliferation, cell-cycle regulation and apoptosis. Histone deacetylase (HDAC) inhibitors have become promising anticancer agents in recent years. They have shown ability to block angiogenesis and cell cycling, as well as initiate differentiation and apoptosis.HDAC inhibition has recently been clinically validated as a new therapeutic strategy for cancer treatment with the FDA approval of suberoylanilide hydroxamic acid (SAHA) (1) for the treatment of cutaneous T cell lymphoma.
(1)
Thus, HDACs are among the most promising therapeutic targets for cancer treatment, and they have inspired researchers to study and develop HDAC inhibitors.
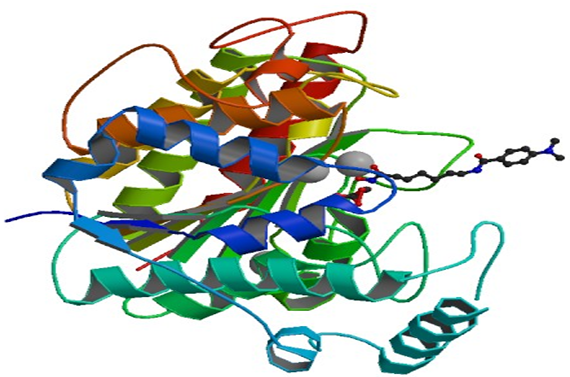
Fig 1.Crystal Structure of HDAC8 Complexed with M344
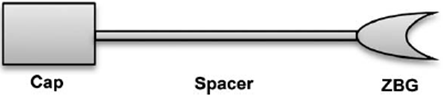
Fig 2. HDACIs Common Pharmacophore Schematic Segmentation.
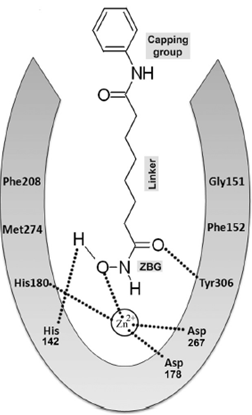
Fig 3. Schematic Representation of Basic Pharmacophore Model and Binding of Hydroxamic acid based HDAC Inhibitors.
OBJECTIVES
In a prototypical HDAC inhibitor, the cap group could be linked to the aliphatic linker group through either hydrogen-bond accepting or donating groups such as keto- and amide-groups. The apparent lack of preference for H-bond donor or acceptor in the linking-moiety presents an opportunity to incorporate other more synthetically accessible and pharmacokinetically desirable moieties that may help simplify the molecular design and synthesis of novel HDAC inhibitors. We proposed that a piperazine ring could act as a linking moiety which joins the cap group to the linker group in a HDAC inhibitor.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to PharmaTutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
EXPERIMENTAL WORKS
Materials & method:
* Operating System : LINUX Fedora -5.
* Softwares : Autodock 1.4.6, Chemsketch, open-babel, chimera.
* Server used : rcsb.org, Dundee prodrg server,
PROCEDURE:
Target (histone deacetylase) Preparation:
· The desired target i.e, histone deacetylase with the entry code 3MZ4 ,was procured from rcsb.org.
· For clarification purpose we have sliced the B chain and the ligand i.e.; B3N501 leaving behind the A chain where the ligand was previously placed.
· All hydrogens were added to the remaining part of the enzyme.
· The newly prepared enzyme was further saved into pdb format.
Ligand Preparation:
· A series of 5 molecules with structural similarity were sketched in chemsketch. Each molecule was further copied for introducing into PRODRG window. After fulfilling the requirement given by PRODRG the AUTODOCK compatible ligand was generated.
· The necessary informations obtained from PRODRG were copied and pasted into notepad, saved as lig.pdb.
· Number of torsions were identified followed by conversion of atom type were made through AD4 type command, subsequently charges were assigned.
· The ligand thus obtained was further saved as lig.pdbqt format.
Preparation of glg (grid log file) file :
· Flexible residue (HIS142) was identified
· Atoms were made AD4 type and charges were assigned.
· Number of torsions were chosen from the selected flexible residue.
· Later two different pdbqt files were prepared,
· i. receptor_flex.pdbqt (when amide torsions were allowed and
· ii. receptor_rigid.pdbqt (when amide torsions were not allowed)
· receptor_rigid.pdbqt & lig.pdbqt were opened respectively in the grid then size of the box changed (usually it has been assigned 100, 100, 100)for proper exploration of the receptor.
· Then it was saved as .gpf format.
· Autogrid4 program was run & the corresponding glg file was prepared.
Preparation of dlg file :
· Through the Docking options, all the required files were opened such as rigid file, flexible file & the ligand.
· From the search parameters genetic algorithm window was opened, depending upon the need all the alteration have been made like number of GA runs, etc and subsequently it was closed.
· Then, docking parameter option was toggled on & saved as .dpf format.
· Autodock4 program was run & the corresponding dlg file was prepared.
Docking result analysis:
Following commands were used, while analyzing the docking output:
· Dlg file - analyse - load conformations - play .
· Open - macromolecule (rigid file name).
· Open - grid & set OA map, Click to off the line as well as show box-close the widget & reduce the iso value to minimize the unwanted crowding.
· Previous macromolecule was replaced.
· Display msms & close contacts are closed for proper visualization & hide the rigid receptor file was hidden, msms option was opened.
· Close contacts was closed for clarification purpose.
RESULTS & DISCUSSION:
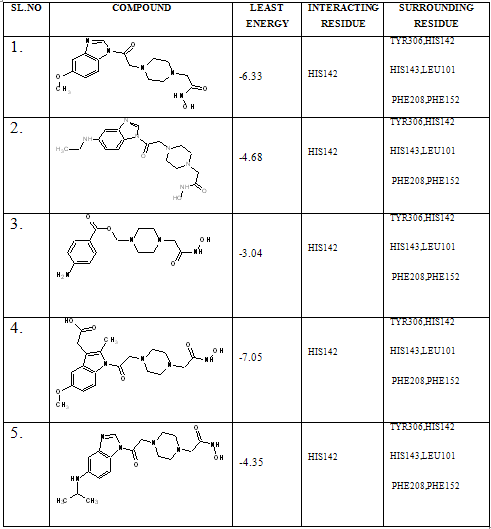
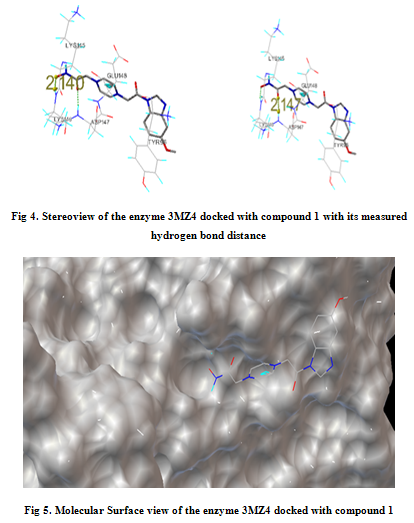
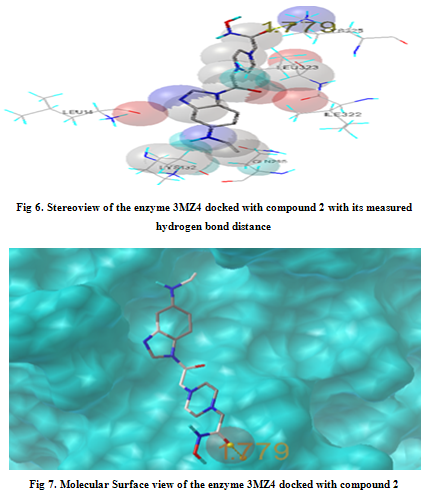
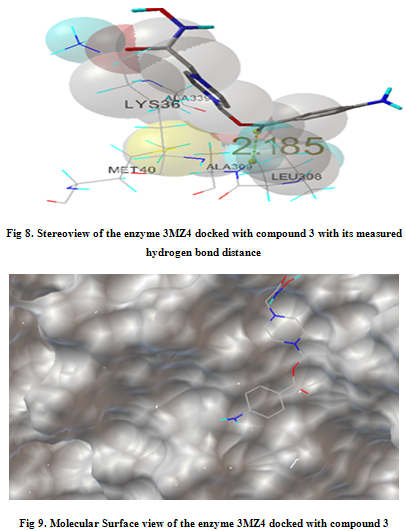
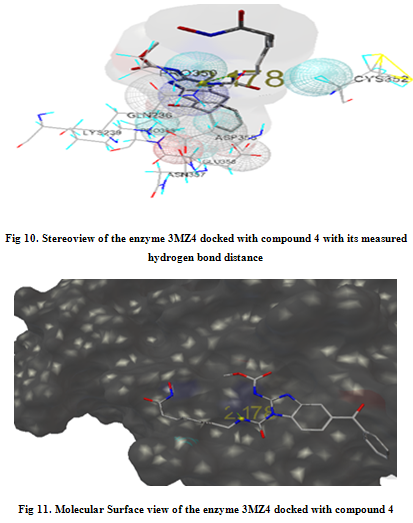
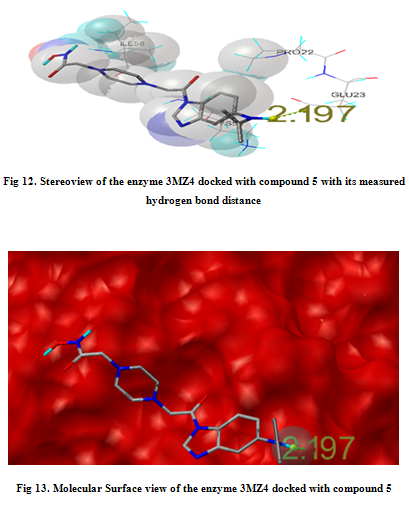
In our present work, we have tried to develop computationally few HDAC inhibitors as novel anticancer drugs. The docking study of each molecule was carried out with 100 number of GA runs. For each molecule 100 different conformers were generated. The binding energy of the least energy conformer was documented in table 1 along with their interacting and surrounding residues respectively. While analyzing the surface topology of the enzyme, with the help of CHIMERA have chosen the flexible residue as HIS142. The space occupied by the grid box was kept 100*100*100 for x, y, z axis respectively.
The scoring functions have been shown in the table to follow. The conventional design considering the literature review were strongly supported by the graphical represention as given in the next part to follow. Hydrogen bonding interaction have been found in all the cases, which also affirms the probable effectiveness of the molecule under study.
Table 1. Details of Docking Output of the compounds (1-5)
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to PharmaTutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
CLOSING REMARK
As it is a preliminary approach in designing molecules which may have an effective therapeutic potentials as HDAC inhibitors nevertheless, the docking results offered us much useful information to understand the interaction mode between such inhibitors and protein and will facilitate the design of more potent inhibitors.
REFERENCE
1.Bai, S. Ghoshal, K. Mol. Cell. Biol. 2005, 25, 751–66.
2.Bouras, T. Fu, M. J. Biol. Chem. 2005, 280, 10264–76.
3.Bradbury, C. A. Khanim, F. L. Hayden, R. Bunce, C.M. Leukemia, 2005, 19, 1751–59.
4.Brehm, A. Miska, E. A. McCance, D. J. Nature, 1998, 391, 597–601.
5.Chen, W. Y. Wang, D. H. Cell, 2005, 123, 437–48.
6.Kawai, H. Li, H. Int. J. Cancer, 2003, 107, 353–58.
7.Kelly W.K. Marks, P. A. Nat. Clin. Pract. Oncol, 2005, 2, 150–57.
8.Monneret, C. Histone deacetylase inhibitors, Eur. J. Med. Chem. 2005, 40, 1–13.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to PharmaTutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE









