About Authors:
Emanual Michael Patelia*, Rakesh Thakur, Jayesh Patel
Department of Pharmacology,
University of Bedfordshire, Luton, LU1 3JU, England.
*ricky.emanual@gmail.com
Abstract
The PCR is widely used technique in sex determination. PCR is also used for the determination of x linked inherited disorders, with help of biopsied embryos into mothers. DNA polymerase enzyme replicates a piece of DNA. Thus, chain reaction occurs and generating multiple copies of it. Polymerase chain reaction is frequently used technique for sex determination. PCR can also be used for detecting the presence or absence of a particular piece of DNA. In this method, we used PCR method for determination of the sex of three unknown bovine samples.PCR techniques have developed to reduce the problems by increasing amplification quality.
REFERENCE ID: PHARMATUTOR-ART-1911
Introduction:
Polymerase chain reaction is one type of the method used for the determination of the sex. Polymerase chain reaction method (PCR) was invented by Kary B. Mullis, they was also received the Nobel prize in chemistry for PCR method invention on 10 December, 1993. [1] PCR Method also used in other detection like mutations detecting, monitoring cancer therapy, detecting bacterial, viral infection and also sex determination. [1]
Sex determination is one of the important methods, which help to determine the sex of the developing embryo. Mostly x-y system is use for the sex determination in human and mammals. Males have ‘x’ chromosome (xy) and female have both (xx) chromosome. So with the help of the chromosome sex are determine. It is not true for all the organism, for example in bird and reptiles male have homogametic (ZZ) and female have heterogametic (ZW). Generally in male x and y chromosome and in female xx chromosome. sex is determined by the presence and absence of the y chromosome.[2]
We can determine sex with the help of presence of unique sequence on the y chromosome (SRY gene). The presence of SRY gene and protein encoded is very important for testis differentiation in the early embryo. if SRY gene is absent means development of ovaries. The protein of high mobility group (HMG) family is encoded with the help of SRY gene. After that this protein triggers all the step which is required to development of male phenotype.
Material and Methods:
DNA purification:
In the multiplex PCR method , In these unknown samples adding buffer AL ,BUFFER AW1 and buffer AW2 for isolation and purification of DNA from unknown sample. After addition to each given sample followed centrifugation.
Than transfer the column into new 1.5 ml tube than add buffer AE and incubated at room temperature and than centrifuge. After that discard the column and leave the tube on ice for PCR set up.
PCR:
In these PCR method we used different reagent for the PCR reaction like Buffer, primers,dNTPs, water and Taq DNA polymerase. First we marked the label on six 0.2 ml tube as a ‘M’, ‘F’, ‘1’, ‘2’ , ‘3’ and ‘-‘ .
Than we prepared a master mix in tube by added 15µl PCR buffer,dNTP mix 2.4 µl,primer mix 15 µl, sterile PCR water 103.2 µl,Taq DNA polymerase 2.4 µl . Finally, we prepared a 138µl master mix.
After that we aliquot 23 µl of the master mix into 5 remaining tubes and added 2 µl DNA to each tube. Than placed in the thermo cycler. In the last PCR thermal cycle keep for initial denaturation at 95 °C for 1 min, annealing at 66 °C for 1 min, extension at 72°C and final extension at 72°C for 3 min. Than for remaining use stored the samples at -20°C.
Gel Electrophoresis:
First Place the 16 sample comb in the MIDI Gel electrophoresis. After that we poured gel solution (1.5% agarose) to a depth of 5mm and allow to set for 15 min.
Than we add 5 µl of sample loading buffer to each o.2ml tube from the PCR reaction.
When the gel has set. Take the comb and add electrophoresis buffer (TAE) to cover the gel by approximately 0.5cm.add 20 µl of all sample to individual well and than run the gel at 100v until the blue dye runs half way down the gel.
DNA quantification:
DNA quantification are help to determine the quantity of amplifiable DNA [3]. Quantities analysis is a procedure where the concentration of a particular bimolecule in a sample is determined [4].with the help of a spectrophotometer the DNA and proteins are determine by absorbance of uv light. At 260nm absorbance DNA determined and 280nm absorbance protein contamination was estimated. The blank sample was used before measuring each sample absorbance.
Results:
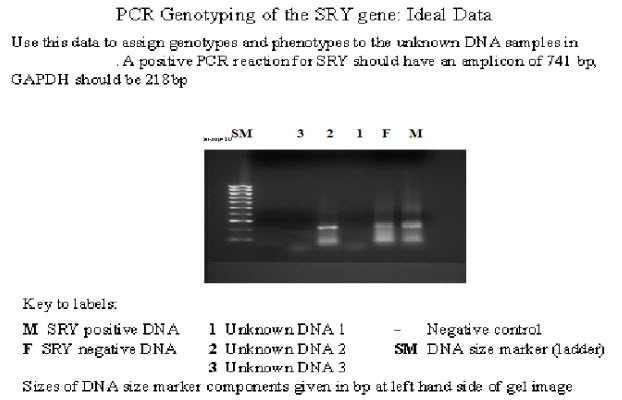
Figure 1: A positive PCR reaction for SRY should have an amplicon of 741bp, GAPDH should be 218bp.
The first lane in the figure above is marker sample.
second lane is male
Third lane is female
Forth lane is sample 1
Fifth lane is sample 2
Six lane is unknown sample 3
Seven lane is negative control
In the second lane, it is for male control. Ideally two bands should be detected. One bands at the level of 218bp and second band at 741bp, First band for GAPDH gene and second band for SRY gene. In our experiment result two week bands are visible.
Third lane is for female. There is only one band are visible at 218 bp , which is for GAPDH gene. In female there is no SRY gene. In our experiment we detected week band at 218bp in second lane.
Forth lane is for unknown sample 1. This sample was female, there visible only one band at 218bp and there is no SRY gene visible. In our experiment one band is visible at 218 and no other band are visible.
Five lane is for unknown sample 2. This sample was male, there should two bands are visible, one band at 218bp for housekeeping GAPDH gene and second band at 741bp level for SRY gene. In our experiment one week band at 218bp and no other band is visible.
Six lane is for unknown sample 3. This sample was male. there should two bands are visible, one band at 218bp for housekeeping GAPDH gene and second band at 741bp level for SRY gene. In our experiment two bands are visible; first band at 218bp for GAPDH and second week bands at 741bp for SRY gene.
Seven lane for negative control. There should be no band are visible. In our experiment bright band is shown, which is some similar to the primer dimer.
1. The terms regarding denaturing, annealing and extension refer to in PCR.
Denaturing means heating of the PCR products upto 94 °C for 30 sec .because of these hydrogen bonds between complementary bases is broken and get single stranded DNA molecules.
Annealing means lowering the temperature at 66°C for 1 min which are help in the binding of primers to single-stranded DNA templates.
Extension means heating of mixture to 72°C for 1 min to which helps to DNA polymerase to synthesize a new DNA strand complementary to DNA template.
2. Explanation regarding the role of GAPDH primers in this experiment.
GAPDH genes are housekeeping genes. GAPDH primers were provided to amplify these genes.
3. Explanation regarding the role of SRY primers in this experiment.
SRY gene which is sex determining region on Y Chromosome. For differentiation of testis in human embryo, the presence of SRY gene and the protein encoded by it are necessary. If SRY gene is absent, it results in development of ovaries SRY primers were provided to amplify these genes.
4. Calculation of GC content of each primer. Show your workings.
GC content= G+C/ A+T+G+C × 100
GAPDH F: 12/12+12 × 100 = 50
GAPDH R: 12/12+12 × 100 = 50
SRY F: 11/13+11 × 100 = 45.83
SRY R: 11/13+11 × 100 = 45.83
5. Calculation of the volume of each reagent in the master mix that will be present in each PCR tube. Show your working.
PCR buffer: 15/6 = 2.5
Dntp mix: 2.4/6 = 0.4
Primer mix: 15/ 6 = 2.5
Taq DNA polymerase: 2.4/6 = 0.4
6. Calculation for the final concentration of dNTPs present in each PCR tube. Show your workings.
C1V1 =C2V2
50 Mm × 0.4 µL = C2 × 25 µL
C2 = 0.8 Mm
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE
Table 1: List of components used for the calculations.
|
|
DNA 4a |
DNA 4b |
DNA 4c |
|
Abs 260 |
0.081 |
0.496 |
0.205 |
|
Abs 280 |
0.043 |
0.603 |
0.238 |
|
Ratio Abs 260/Abs 280 |
1.883 |
0.822 |
0.861 |
|
DNA concentration in the cuvette (µg DNA ml) |
4.05 |
24.8 |
10.25 |
|
The dilution factor for the sample |
49 |
49 |
49 |
|
DNA concentration in original sample (µg DNA ml) |
198.45 |
1215.2 |
502.25 |
|
The amount of DNA in 20 µl sample (µg) |
3.969 |
24.304 |
10.045 |
7. Which DNA sample has the highest concentration?
DNA 4b has the highest concentration (1215.2)
8. Which DNA sample is the most pure?
DNA 4a is the purest DNA with ratio of 1.883
9. Which DNA sample is the most suitable for use in PCR?
DNA4a
Discussion
There should be many errors possible at the time of the experiment like at the time of the addition of the reagents, some reagent miss in the master mix and some time possible errors with the concentration of the reagent.
Sometime large concentration of the magnesium also affect to the DNA and presence of the protein.so we can not get a specific amplification. [5]
Co-solvent and other additive like dimethyl sulfoxide and formamide also affect to the polymarase chain reaction. .[5]
In the performing of the PCR,primer designing programs are very important [6]. Because of not proper designed prime can not get the proper result. (You et al., 2008)
The most important affecting factor is selection of proper primers, which affecting the polymerase chain reaction [8]. For the specific amplification specific primers are requires otherwise do not match to other target in certain orientations and in some area that allow undesired amplification [8].
Sometime because of the not proper interaction between the primer and template efficiently errors occurs and also some contamination affect in the PCR reaction.
200-500bp is ideal regions for amplification for analytical purpose [1]. Amplification should be difficult to detect on agarose gel if region smaller than 200bp and amplification should be not proper if strength is higher [1]. The sequence of the primer is also very important [1].
Two primers same in base composition and in length, it means two primers should be similar annealing temperatures.
There are some factor which are affect the design of the primers. Like oligonucleotide made sequence with complementary itself or other primer, because of these base pairing even within the primers or pairing between the primers, instead of other target sequence [1]. Because of presence of large quantity of the primers, they encounter a partially complementary molecule, rather than a perfectly complementary template molecule, forming pairing .when binding are done produce a fragment known as primer dimmers [1].
Role of SRY gene in gonadal development
Mammalian embryos have bipotential gonads, base on the paternal and maternal chromosomal presence they can develop either into a testis or an ovary [7]. Contribution of the x chromosome result in ovarian differentiation and the y chromosome result in testicular differentiation. At the time of development, bipotential gonad contain a pools of undifferentiated somatic cell precursors, which differentiate into either testicular or ovarian [7].
On y chromosome the gene for the transcription factory SRY is located and it is responsible for take fate of testis in bipotential gonad. There is SF1 factor (steroidogenic factor) called nuclear orphan receptor. Which regulate immediate downstream gene called as SOX9 [7]. It is done with the help of the interaction on its testis specific enhance. The role of the SOX9 is to contro differentiation of precursor cell into Sertoli cells. FGF9 and prostaglandins with the help of SOX9 promote testis development.
The process of the sex determination start with differentiation of somantic cells. The sequence on the DNA is observed when binding of SRY to HMG box[7]. Helix loop helix (bHLH) transcription factors have identified by scientists and they are known to monitor cell fate specification, morphogenesis and differentiation. Early development of embryo bHLH gene play some role and affect a wide variety of tissues [7]. tcf21 is a one type of gene which help in encodes transcription factor for family bHLH. During male gonadal sex determination SRY gene interacts with Tcf21,it is hypothesized and promotes somatic cell differentiation
The polymerase chain reaction is one of the most important technique in determining sex of given sample.PCR is very useful for study and the analysis of genes [8].
References
1)Dale, J. von Schantz, M. and Plant, N. (2011). From Genes to Genomes: Concepts and Applications of DNA Technology, pp. 400. Chichester, West Sussex: John Wiley & Sons.
2)Khana,Pragya.(2008) cell and molecular biology,pp.232
3)Butler, J. M. (2005). Forensic DNA Typing [Electronic Resource]: Biology, Technology, and Genetics of STR Markers, pp. 660. London: Elsevier Academic Press.
4)Langford, A. (2005). Practical Skills in Forensic Science, pp. 516. Harlow, England ; New York: Pearson Prentice Hall.
5)Seviour,R.J,Nielsen,Per.H .(2008).Microbial Ecology Of Activated Sludge,pp.354
6)You, F. M., Huo, N., Yong, Q. G., Ming-cheng Luo, Ma, Y., Hane, D., Lazo, G. R., Dvorak, J. and Anderson, O. D. (2008). BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinformatics 9, 1-13.
7)Bhandari , R. K., Sadler-Riggleman, I., Clement, T. M. and Skinner, M. K. (2011). Basic helix-loop-helix transcription factor TCF21 is a downstream target of the male sex determining gene SRY. PLOS ONE 6, 1-11. 8)Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S. and Madden, T. L. (2012). Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134-144.
NOW YOU CAN ALSO PUBLISH YOUR ARTICLE ONLINE.
SUBMIT YOUR ARTICLE/PROJECT AT articles@pharmatutor.org
Subscribe to Pharmatutor Alerts by Email
FIND OUT MORE ARTICLES AT OUR DATABASE











.png)

